Type-1 Self Incompatibility (SI)
Overview
| Family | Solanaceae |
| Genus | 98 (16 assembled genus) |
| Species | 2700 (116 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SLFs (pollen specific expression) |
Organism Image

Description
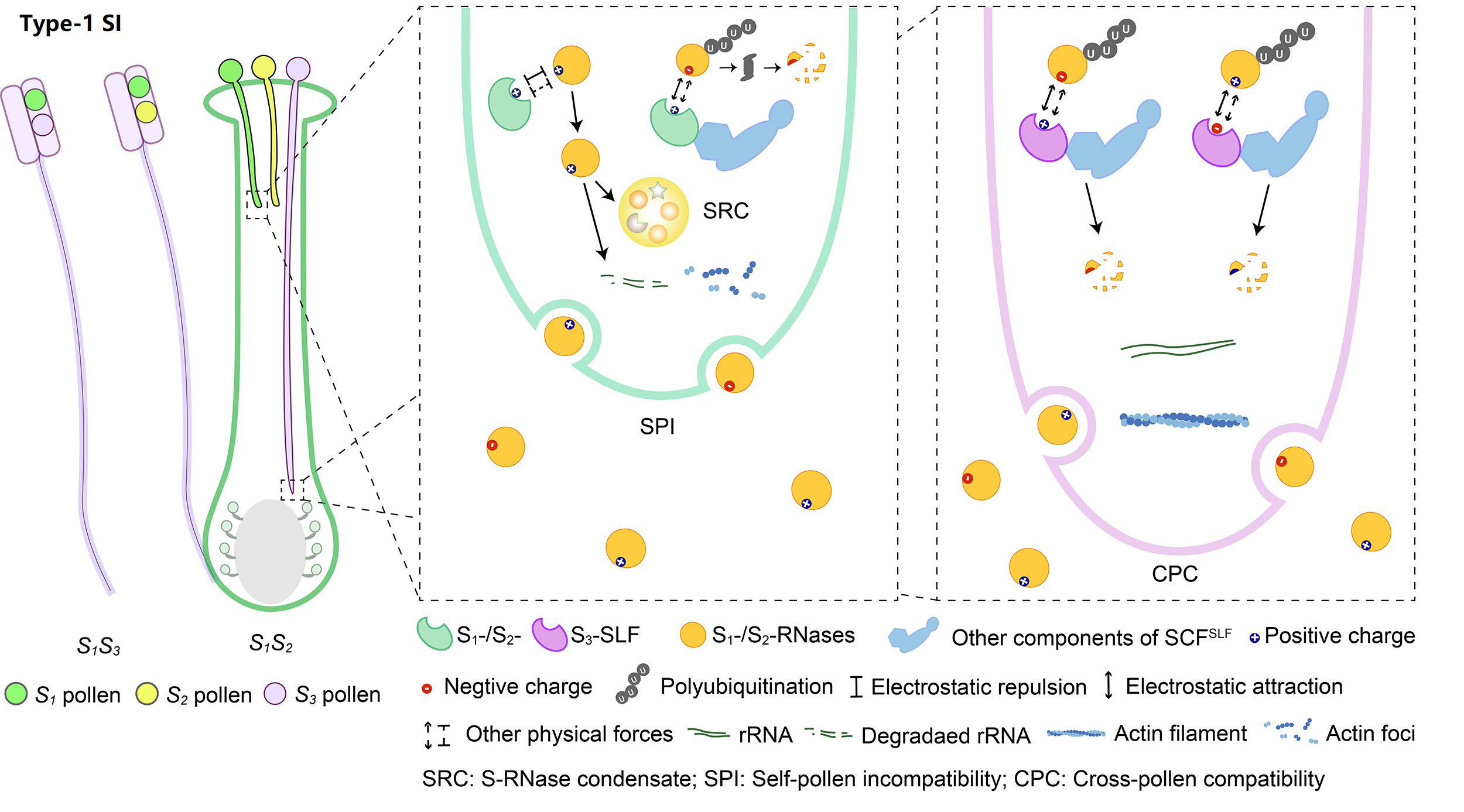
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.
SI genes
Publication
Anderson, M., Cornish, E., Mau, SL. et al. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321, 38–44 (1986). https://doi.org/10.1038/321038a0.
Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004 May 20;429(6989):302-5. doi: 10.1038/nature02523.
Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, Kao TH, Takayama S. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science. 2010 Nov 5;330(6005):796-9. doi: 10.1126/science.1195243.
Liu W, Fan J, Li J, Song Y, Li Q, Zhang Y, Xue Y. SCF(SLF)-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida. Front Genet. 2014 Jul 22;5:228. doi: 10.3389/fgene.2014.00228.
Li J, Zhang Y, Song Y, Zhang H, Fan J, Li Q, Zhang D, Xue Y. Electrostatic potentials of the S-locus F-box proteins contribute to the pollen S specificity in self-incompatibility in Petunia hybrida. Plant J. 2017 Jan;89(1):45-57. doi: 10.1111/tpj.13318.
Zhao H, Song Y, Li J, Zhang Y, Huang H, Li Q, Zhang Y, Xue Y. Primary restriction of S-RNase cytotoxicity by a stepwise ubiquitination and degradation pathway in Petunia hybrida. New Phytol. 2021 Aug;231(3):1249-1264. doi: 10.1111/nph.17438.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Tian H, Zhang H, Huang H, Zhang Y, Xue Y. Phase separation of S-RNase promotes self-incompatibility in Petunia hybrida. J Integr Plant Biol. 2024 May;66(5):986-1006. doi: 10.1111/jipb.13584.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Overview
| Family | Plantaginaceae |
| Genus | 107 (7 assembled genus) |
| Species | 1900 (10 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SLFs (pollen specific expression) |
Organism Image

Description
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.

SI genes
Publication
Xue Y, Carpenter R, Dickinson HG, Coen ES. Origin of allelic diversity in antirrhinum S locus RNases. Plant Cell. 1996 May;8(5):805-14. doi: 10.1105/tpc.8.5.805.
Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol. 2002 Sep;50(1):29-42. doi: 10.1023/a:1016050018779.
Qiao H, Wang F, Zhao L, Zhou J, Lai Z, Zhang Y, Robbins TP, Xue Y. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell. 2004 Sep;16(9):2307-22. doi: 10.1105/tpc.104.024919.
Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y. The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell. 2004 Mar;16(3):582-95. doi: 10.1105/tpc.017673.
Zhao L, Huang J, Zhao Z, Li Q, Sims TL, Xue Y. The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J. 2010 Apr 1;62(1):52-63. doi: 10.1111/j.1365-313X.2010.04123.x.
Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, Kao TH, Takayama S. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science. 2010 Nov 5;330(6005):796-9. doi: 10.1126/science.1195243.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview
| Family | Rosaceae |
| Genus | 91 (19 assembled genus) |
| Species | 4828 (74 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SFB/SFBB (pollen specific expression) |
Organism Image

Description
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.

SI genes
Publication
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H. Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet. 1996 Mar 20;250(5):547-57. doi: 10.1007/BF02174443. Erratum in: Mol Gen Genet. 1996 Aug 27;252(1-2):222. doi: 10.1007/BF02173225.
Ishimizu T, Shinkawa T, Sakiyama F, Norioka S. Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Mol Biol. 1998 Aug;37(6):931-41. doi: 10.1023/a:1006078500664.
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell. 2003 Mar;15(3):771-81. doi: 10.1105/tpc.009290.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview
| Family | Rutaceae |
| Genus | 160 (8 assembled genus) |
| Species | 1600 (24 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SLFs (pollen specific expression) |
Organism Image

Description
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.

SI genes
Publication
Liang M, Cao Z, Zhu A, Liu Y, Tao M, Yang H, Xu Q Jr, Wang S, Liu J, Li Y, Chen C, Xie Z, Deng C, Ye J, Guo W, Xu Q, Xia R, Larkin RM, Deng X, Bosch M, Franklin-Tong VE, Chai L. Evolution of self-compatibility by a mutant Sm-RNase in citrus. Nat Plants. 2020 Feb;6(2):131-142. doi: 10.1038/s41477-020-0597-3.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.