Self Incompatibility (SI) Type
Overview (Solanaceae)
| Family | Solanaceae |
| Genus | 98 (16 assembled genus) |
| Species | 2700 (116 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SLFs (pollen specific expression) |
Organism Image

Description (Solanaceae)
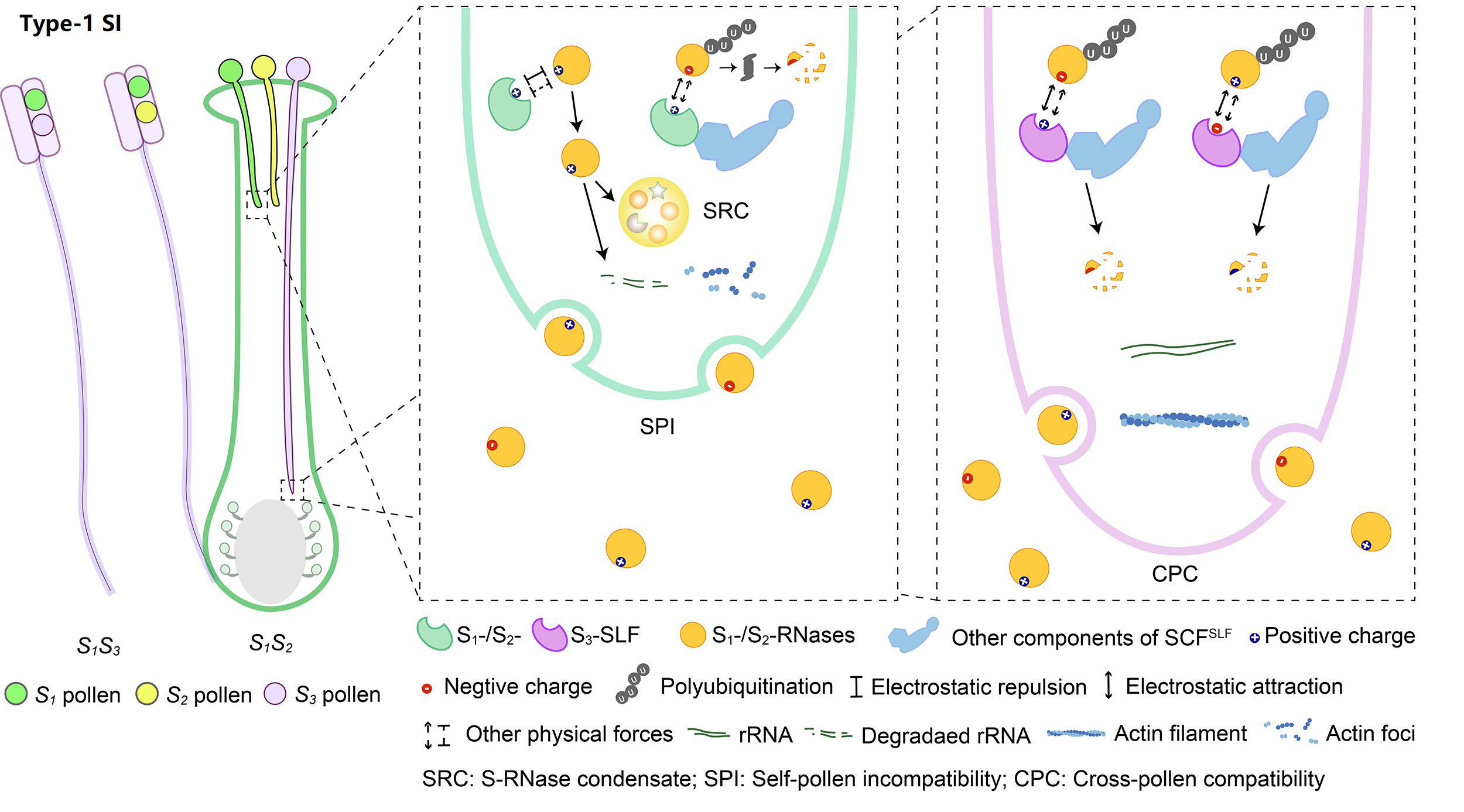
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.
Publication (Solanaceae)
Anderson, M., Cornish, E., Mau, SL. et al. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321, 38–44 (1986). https://doi.org/10.1038/321038a0.
Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004 May 20;429(6989):302-5. doi: 10.1038/nature02523.
Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, Kao TH, Takayama S. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science. 2010 Nov 5;330(6005):796-9. doi: 10.1126/science.1195243.
Liu W, Fan J, Li J, Song Y, Li Q, Zhang Y, Xue Y. SCF(SLF)-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida. Front Genet. 2014 Jul 22;5:228. doi: 10.3389/fgene.2014.00228.
Li J, Zhang Y, Song Y, Zhang H, Fan J, Li Q, Zhang D, Xue Y. Electrostatic potentials of the S-locus F-box proteins contribute to the pollen S specificity in self-incompatibility in Petunia hybrida. Plant J. 2017 Jan;89(1):45-57. doi: 10.1111/tpj.13318.
Zhao H, Song Y, Li J, Zhang Y, Huang H, Li Q, Zhang Y, Xue Y. Primary restriction of S-RNase cytotoxicity by a stepwise ubiquitination and degradation pathway in Petunia hybrida. New Phytol. 2021 Aug;231(3):1249-1264. doi: 10.1111/nph.17438.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Tian H, Zhang H, Huang H, Zhang Y, Xue Y. Phase separation of S-RNase promotes self-incompatibility in Petunia hybrida. J Integr Plant Biol. 2024 May;66(5):986-1006. doi: 10.1111/jipb.13584.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Overview (Plantaginaceae)
| Family | Plantaginaceae |
| Genus | 107 (7 assembled genus) |
| Species | 1900 (10 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SLFs (pollen specific expression) |
Organism Image

Description (Plantaginaceae)
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.

Publication (Plantaginaceae)
Xue Y, Carpenter R, Dickinson HG, Coen ES. Origin of allelic diversity in antirrhinum S locus RNases. Plant Cell. 1996 May;8(5):805-14. doi: 10.1105/tpc.8.5.805.
Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol. 2002 Sep;50(1):29-42. doi: 10.1023/a:1016050018779.
Qiao H, Wang F, Zhao L, Zhou J, Lai Z, Zhang Y, Robbins TP, Xue Y. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell. 2004 Sep;16(9):2307-22. doi: 10.1105/tpc.104.024919.
Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y. The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell. 2004 Mar;16(3):582-95. doi: 10.1105/tpc.017673.
Zhao L, Huang J, Zhao Z, Li Q, Sims TL, Xue Y. The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J. 2010 Apr 1;62(1):52-63. doi: 10.1111/j.1365-313X.2010.04123.x.
Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, Kao TH, Takayama S. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science. 2010 Nov 5;330(6005):796-9. doi: 10.1126/science.1195243.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview (Rosaceae)
| Family | Rosaceae |
| Genus | 91 (19 assembled genus) |
| Species | 4828 (74 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SFB/SFBB (pollen specific expression) |
Organism Image

Description (Rosaceae)
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.

Publication (Rosaceae)
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H. Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet. 1996 Mar 20;250(5):547-57. doi: 10.1007/BF02174443. Erratum in: Mol Gen Genet. 1996 Aug 27;252(1-2):222. doi: 10.1007/BF02173225.
Ishimizu T, Shinkawa T, Sakiyama F, Norioka S. Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Mol Biol. 1998 Aug;37(6):931-41. doi: 10.1023/a:1006078500664.
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell. 2003 Mar;15(3):771-81. doi: 10.1105/tpc.009290.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview (Rutaceae)
| Family | Rutaceae |
| Genus | 160 (8 assembled genus) |
| Species | 1600 (24 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SLFs (pollen specific expression) |
Organism Image

Description (Rutaceae)
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.

Publication (Rutaceae)
Liang M, Cao Z, Zhu A, Liu Y, Tao M, Yang H, Xu Q Jr, Wang S, Liu J, Li Y, Chen C, Xie Z, Deng C, Ye J, Guo W, Xu Q, Xia R, Larkin RM, Deng X, Bosch M, Franklin-Tong VE, Chai L. Evolution of self-compatibility by a mutant Sm-RNase in citrus. Nat Plants. 2020 Feb;6(2):131-142. doi: 10.1038/s41477-020-0597-3.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview
| Family | Brassicaceae |
| Genus | 372 (40 assembled genus) |
| Species | 4060 (79 assembled species) |
| SI type | Type-2 |
| SI genes | SRK (pistil specific expression); SCR/SP11 (pollen specific expression) |
Organism Image

Description
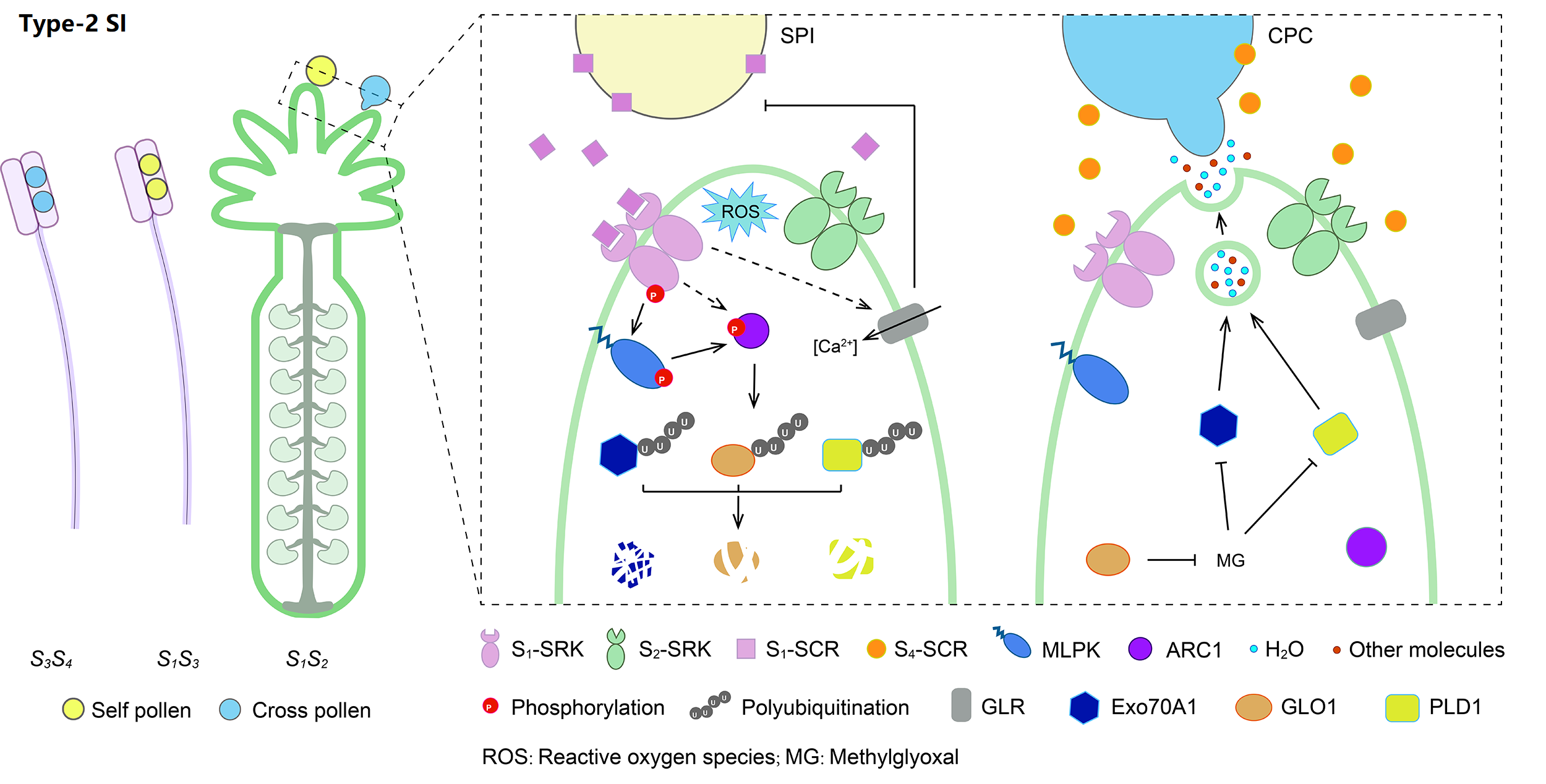
Type-2 SI is the sporophytic Brassicaceae-type SI, controlled by a male S-locus cysteine-rich (SCR) protein/S-locus protein 11 and a female S-locus receptor kinase (SRK). Pollen S1-SCR can specifically recognize its cognate S1-SRK after pollination, triggering several signaling cascades mediated by phosphorylation and ubiquitin-proteasome system, thus leading to self-pollen rejection.
Publication
Takayama S, Shiba H, Iwano M, Shimosato H, Che FS, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A. The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci U S A. 2000 Feb 15;97(4):1920-5. doi: 10.1073/pnas.040556397.
Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature. 2000 Feb 24;403(6772):913-6. doi: 10.1038/35002628.
Sato K, Nishio T, Kimura R, Kusaba M, Suzuki T, Hatakeyama K, Ockendon DJ, Satta Y. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics. 2002 Oct;162(2):931-40. doi: 10.1093/genetics/162.2.931.
Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999 Nov 26;286(5445):1697-700. doi: 10.1126/science.286.5445.1697.
Ma R, Han Z, Hu Z, Lin G, Gong X, Zhang H, Nasrallah JB, Chai J. Structural basis for specific self-incompatibility response in Brassica. Cell Res. 2016 Dec;26(12):1320-1329. doi: 10.1038/cr.2016.129.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview
| Family | Papaveraceae |
| Genus | 42 (5 assembled genus) |
| Species | 775 (13 assembled species) |
| SI type | Type-3 |
| SI genes | PrsS (pistil specific expression); PrpS (pollen specific expression) |
Organism Image

Description
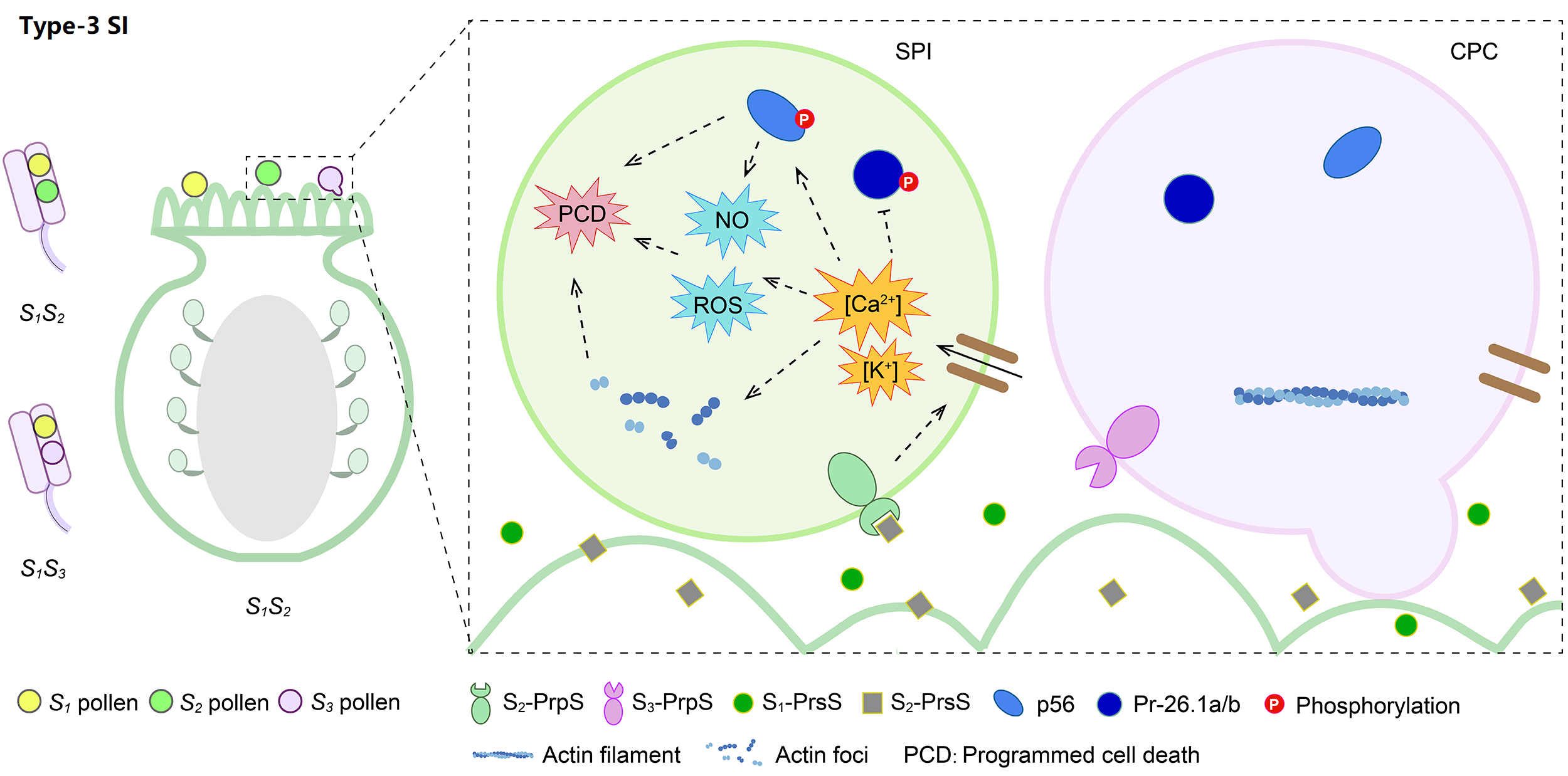
Type-3 is the gametophytic Papaveraceae-type SI, possessing the common poppy (Papaver rhoeas) stigma S (PrsS) and P. rhoeas pollen S (PrpS). Prs S2 secreted by S1S2 papilla cells can specifically bind to pollen membrane-localized Prp S2, stimulating Ca2+ influx, ROS accumulation, actin depolymerization, and PCD of self-pollen.
Publication
Foote HC, Ride JP, Franklin-Tong VE, Walker EA, Lawrence MJ, Franklin FC. Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2265-9. doi: 10.1073/pnas.91.6.2265.
Wheeler MJ, de Graaf BH, Hadjiosif N, Perry RM, Poulter NS, Osman K, Vatovec S, Harper A, Franklin FC, Franklin-Tong VE. Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature. 2009 Jun 18;459(7249):992-5. doi: 10.1038/nature08027IF: 64.8 Q1 . Epub 2009 May 31. Erratum in: Nature. 2016 Mar 3;531(7592):126. doi: 10.1038/nature16181.
Goring DR, Bosch M, Franklin-Tong VE. Contrasting self-recognition rejection systems for self-incompatibility in Brassica and Papaver. Curr Biol. 2023 Jun 5;33(11):R530-R542. doi: 10.1016/j.cub.2023.03.037.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview
| Family | Primulaceae |
| Genus | 22 (2 assembled genus) |
| Species | 1000 (3 assembled species) |
| SI type | Type-4 |
| SI genes | CCM, GLO, CYP, PUM, KFB |
Organism Image

Description
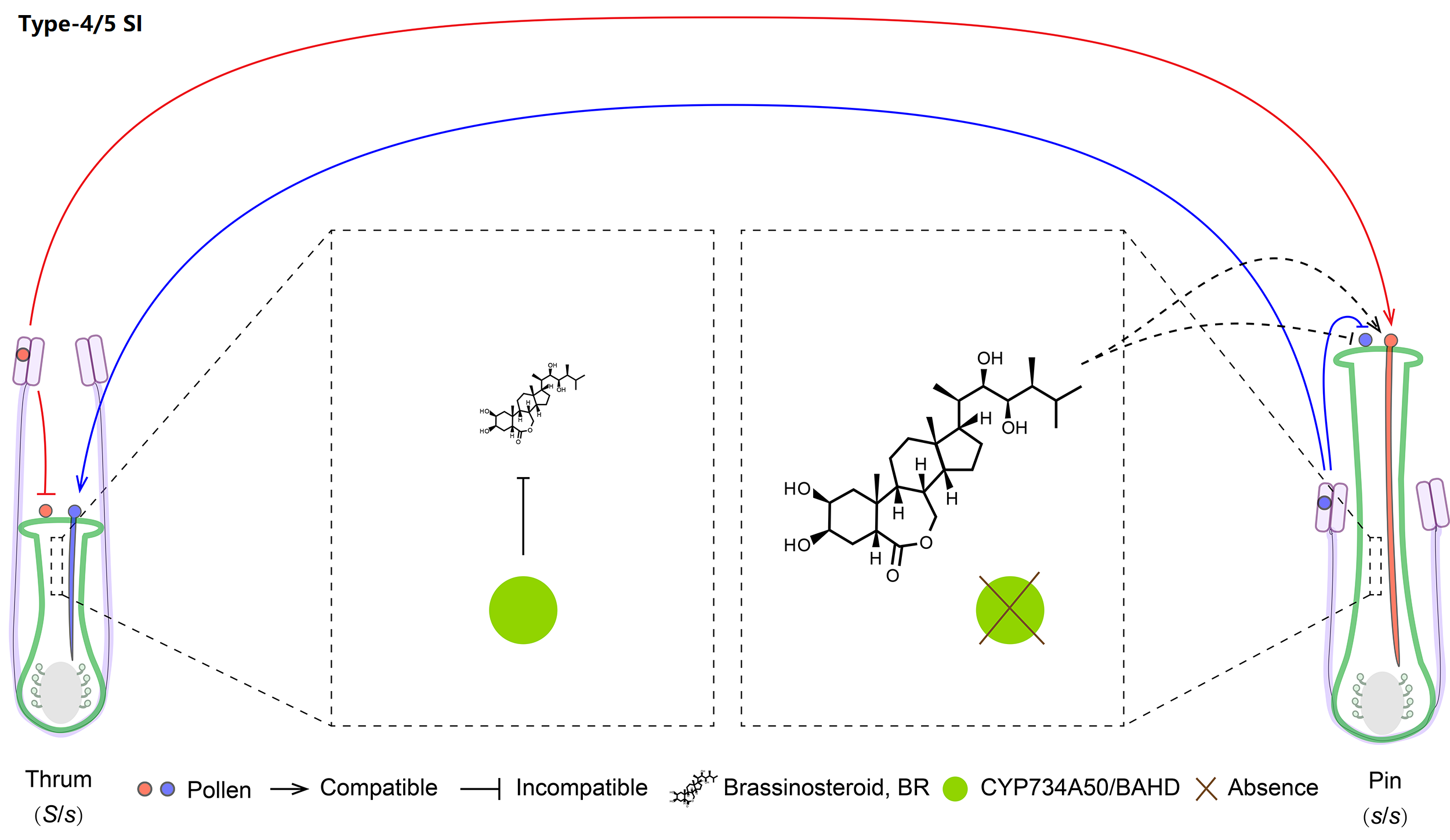
Type-4 and -5 are the sporophytic heterostyly SI of Primulaceae and Turneraceae, controlled by hemizygous S-loci mainly encoding CYP (Cytochrome P450) and TsBAHD, respectively. Athough both of them can inactivate pistil BR, they are absent in the long styles, leading to nonself pollen acceptance and self pollen rejection.
Publication
Li J, Cocker JM, Wright J, Webster MA, McMullan M, Dyer S, Swarbreck D, Caccamo M, Oosterhout CV, Gilmartin PM. Genetic architecture and evolution of the S locus supergene in Primula vulgaris. Nat Plants. 2016 Dec 2;2(12):16188. doi: 10.1038/nplants.2016.188.
Potente G, Léveillé-Bourret É, Yousefi N, Choudhury RR, Keller B, Diop SI, Duijsings D, Pirovano W, Lenhard M, Szövényi P, Conti E. Comparative Genomics Elucidates the Origin of a Supergene Controlling Floral Heteromorphism. Mol Biol Evol. 2022 Feb 3;39(2):msac035. doi: 10.1093/molbev/msac035.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.
Overview
| Family | Passifloraceae |
| Genus | 27 (2 assembled genus) |
| Species | 750 (4 assembled species) |
| SI type | Type-5 |
| SI genes | BAHD, YUC6, SPH1 |
Organism Image

Description
Type-4 and -5 are the sporophytic heterostyly SI of Primulaceae and Turneraceae, controlled by hemizygous S-loci mainly encoding CYP (Cytochrome P450) and TsBAHD, respectively. Athough both of them can inactivate pistil BR, they are absent in the long styles, leading to nonself pollen acceptance and self pollen rejection.

Publication
Shore JS, Hamam HJ, Chafe PDJ, Labonne JDJ, Henning PM, McCubbin AG. The long and short of the S-locus in Turnera (Passifloraceae). New Phytol. 2019 Nov;224(3):1316-1329. doi: 10.1111/nph.15970.
Overview
| Family | Poaceae |
| Genus | 780 (63 assembled genus) |
| Species | 12000 (132 assembled species) |
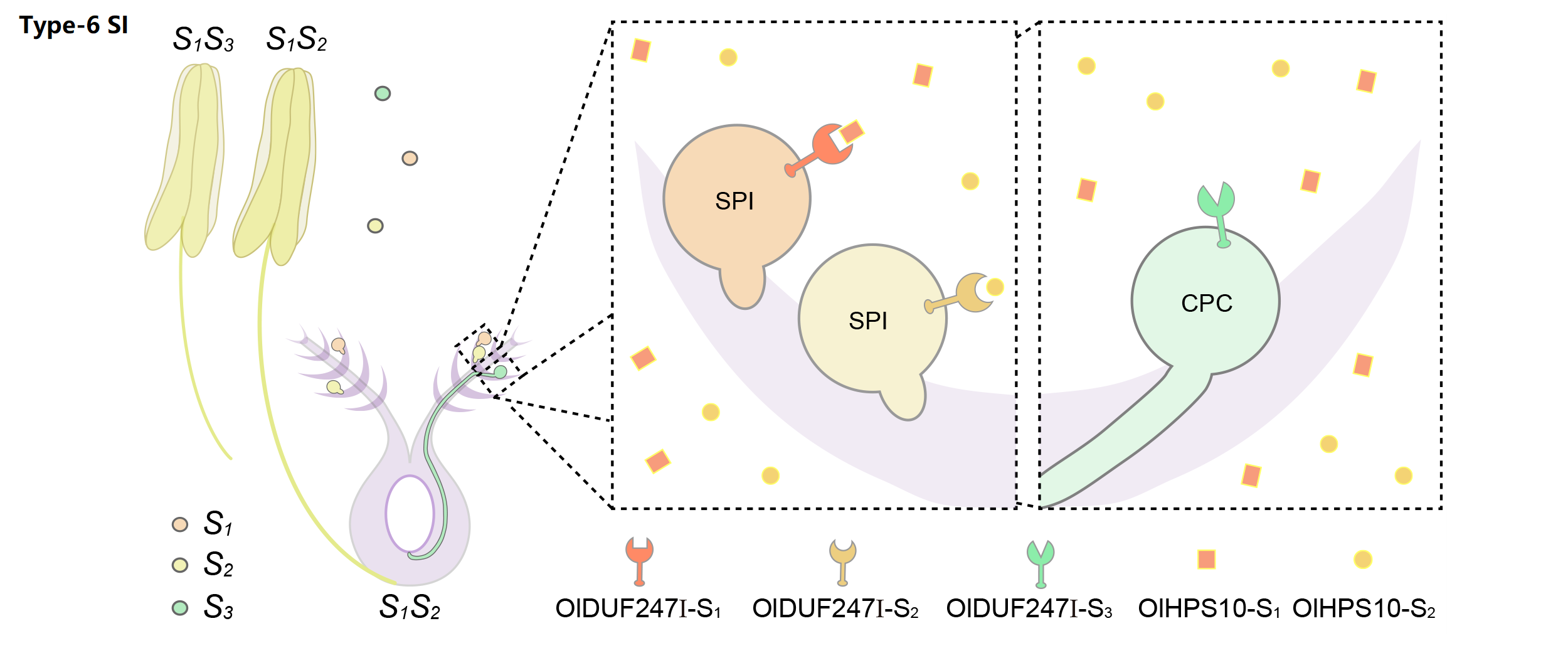
| SI type | Type-6 |
| SI genes | S-loci (HPS10, DUF247), Z-loci (HPS10, DUF247) |
Organism Image

Description
We classified poaceae SI as type-6 gametophytically controlled by two multiallelic and independent loci, S and Z. Taking S as an example, stamen-expressed transmembrane polypeptides containing a domain of unknown function 247 (DUF247) can specifically recognize the pistil-specific peptide HPS10, resulting in the rejection of the self pollen.
Publication
Yanan Wang, Hong Zhao, Sihui Zhu, Hongkui Zhang, Yingying Chen, Xiang Tao, Wenzhi Tong, Huayang Tian, Yu Guan, Huaqiu Huang, Qianqian Han, Zhukuan Cheng, Yijing Zhang, Chuandeng Yi, Yue Zhang, Yongbiao Xue. Control of gametophytic self-incompatibility in the African wild rice, 30 November 2022, PREPRINT (Version 1) available at Research Square https://doi.org/10.21203/rs.3.rs-2121145/v1.
Yang B, Thorogood D, Armstead I, Barth S. How far are we from unravelling self-incompatibility in grasses? New Phytol. 2008;178(4):740-753. doi: 10.1111/j.1469-8137.2008.02421.x.
Manzanares C, Barth S, Thorogood D, Byrne SL, Yates S, Czaban A, Asp T, Yang B, Studer B. A Gene Encoding a DUF247 Domain Protein Cosegregates with the S Self-Incompatibility Locus in Perennial Ryegrass. Mol Biol Evol. 2016 Apr;33(4):870-84. doi: 10.1093/molbev/msv335.
Herridge R, McCourt T, Jacobs JME, Mace P, Brownfield L, Macknight R. Identification of the genes at S and Z reveals the molecular basis and evolution of grass self-incompatibility. Front Plant Sci. 2022 Oct 18;13:1011299. doi: 10.3389/fpls.2022.1011299.
Rohner M, Manzanares C, Yates S, Thorogood D, Copetti D, Lübberstedt T, Asp T, Studer B. Fine-Mapping and Comparative Genomic Analysis Reveal the Gene Composition at the S and Z Self-incompatibility Loci in Grasses. Mol Biol Evol. 2023 Jan 4;40(1):msac259. doi: 10.1093/molbev/msac259.
Overview
| Family | Linaceae |
| Genus | 14 (1 assembled genus) |
| Species | 250 (4 assembled species) |
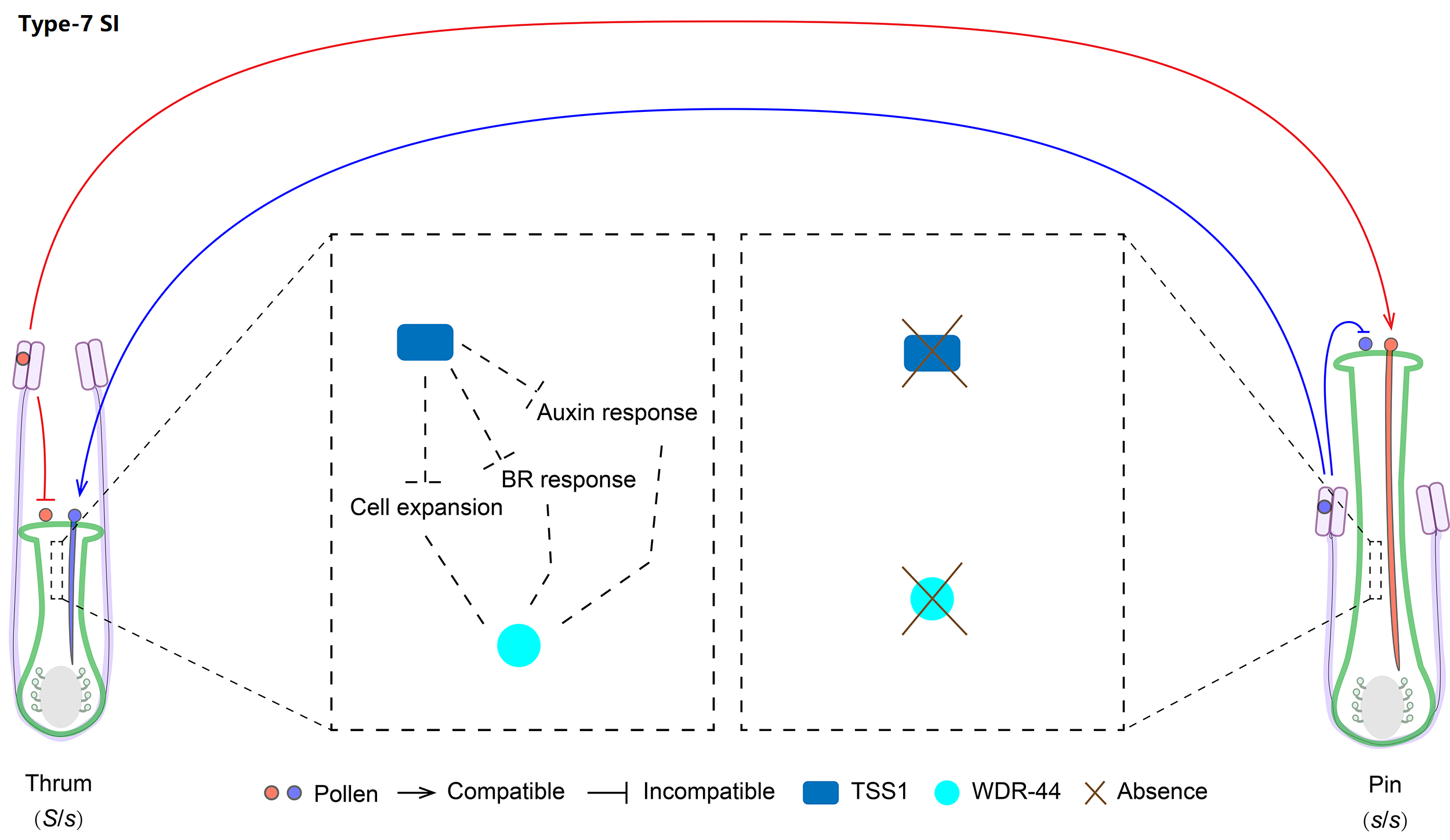
| SI type | Type-7 |
| SI genes | TSS1, WDR-44 |
Organism Image

Description
We classified Linaceae heterostyly SI as type-7 with two distyly candidate genes (LtTSS1 and LtWDR-44) only expressed by the dominant S allele, which can further regulate cell expression, BR and auxin responses of the short styles.
Publication
Gutiérrez-Valencia J, Fracassetti M, Berdan EL, Bunikis I, Soler L, Dainat J, Kutschera VE, Losvik A, Désamoré A, Hughes PW, Foroozani A, Laenen B, Pesquet E, Abdelaziz M, Pettersson OV, Nystedt B, Brennan AC, Arroyo J, Slotte T. Genomic analyses of the Linum distyly supergene reveal convergent evolution at the molecular level. Curr Biol. 2022 Oct 24;32(20):4360-4371.e6. doi: 10.1016/j.cub.2022.08.042.
Overview
| Family | Oleaceae |
| Genus | 28 (8 assembled genus) |
| Species | 700 (12 assembled species) |
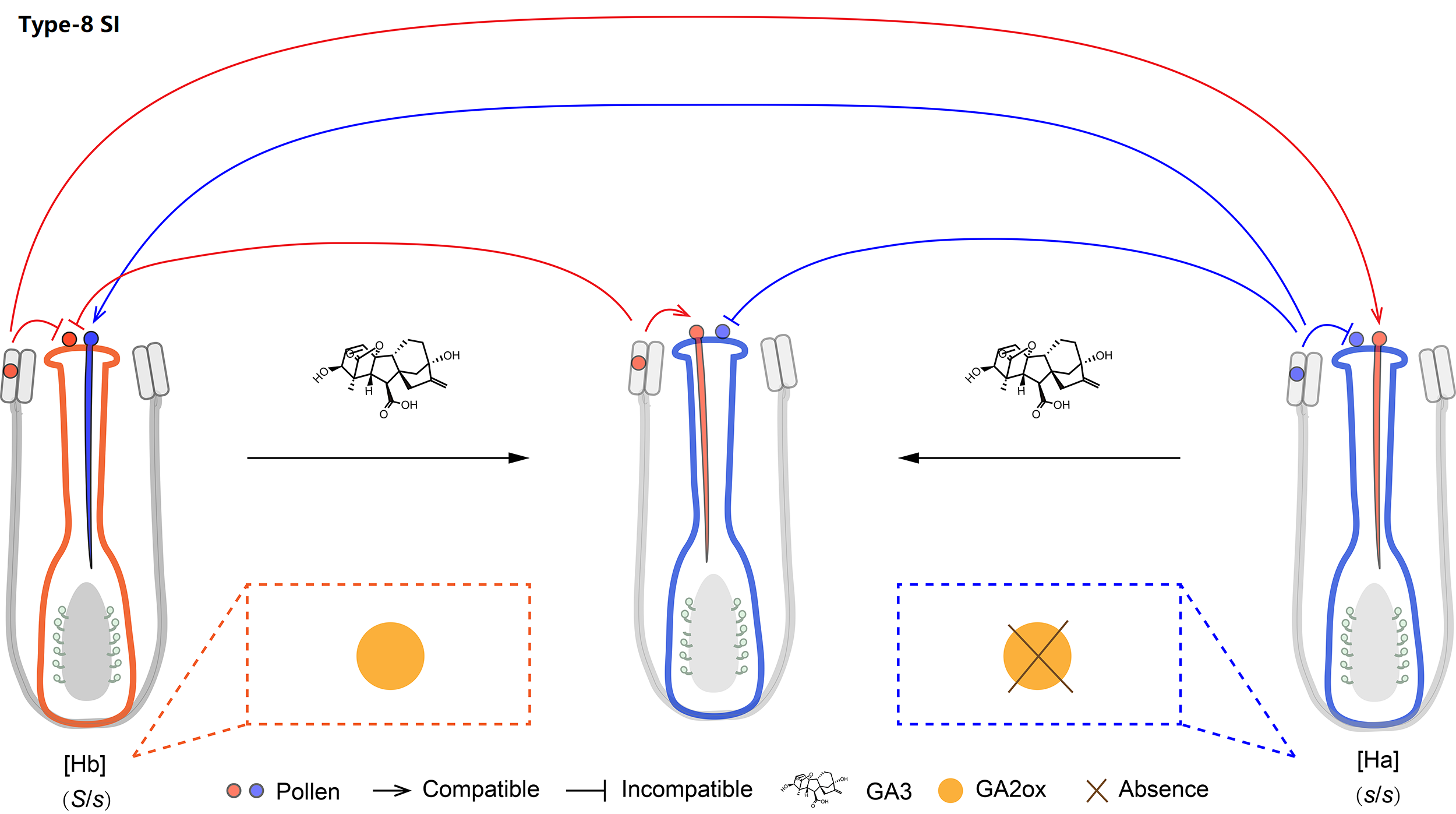
| SI type | Type-8 |
| SI genes | GA2ox |
Organism Image

Description
We classified Oleaceae SI as type-8. The presence/absence polymorphism of the GA2ox-S gene is stably associated with SI groups across Oleaceae. GA3 treatment can switch the female specificity of [Hb] individuals and the male specificity of [Ha] ones.
Publication
Castric V, Batista RA, Carré A, Mousavi S, Mazoyer C, Godé C, Gallina S, Ponitzki C, Theron A, Bellec A, Marande W, Santoni S, Mariotti R, Rubini A, Legrand S, Billiard S, Vekemans X, Vernet P, Saumitou-Laprade P. The homomorphic self-incompatibility system in Oleaceae is controlled by a hemizygous genomic region expressing a gibberellin pathway gene. Curr Biol. 2024 May 6;34(9):1967-1976.e6. doi: 10.1016/j.cub.2024.03.047.