Self Incompatibility (SI) Gene
Type-1 (Solanaceae, Plantaginaceae, Rosaceae, Rutaceae) Supergene, SLF/SFB/SFBB and S-RNase
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil and pollen S components, corresponding to S-RNase and S-locus F-box (SLF), also named S-haplotype-specific F-box (SFB), respectively. So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae, spanning two major clades (super-rosids and superasterids).[ref]
It was postulated that an SLF allelic product specifically detoxifies its non-self S-ribonucleases (S-RNases), allelic products of the pistil S determinant, inside pollen tubes via the ubiquitin–26S-proteasome system, thereby allowing compatible pollinations.[ref]
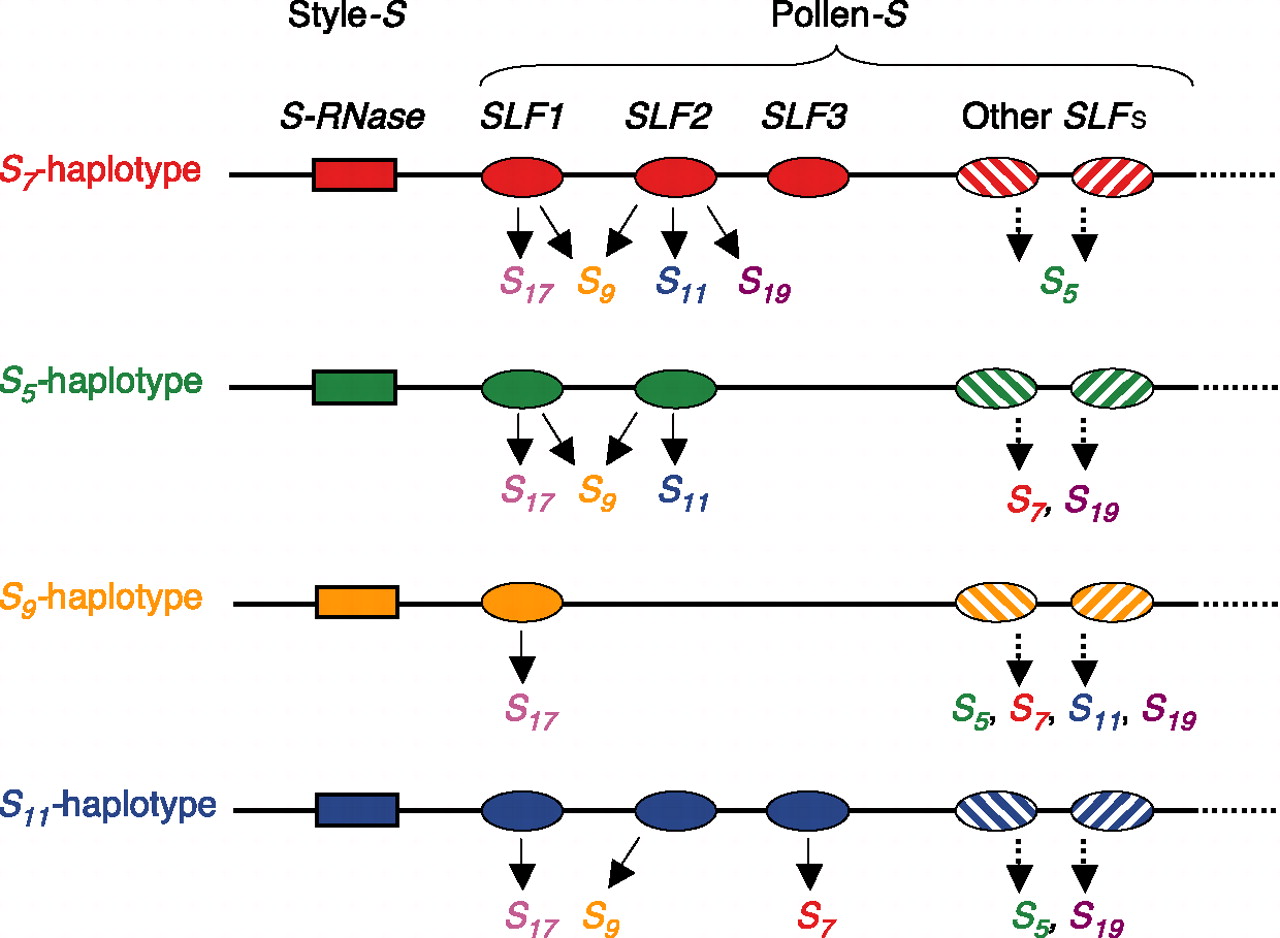
Fig. Collaborative non-self recognition model for SI in Petunia. The single S-RNase gene constituting style-S and the multiple SLFs constituting pollen-S are depicted as boxes and ovals, respectively. The locations of these SLFs relative to S-RNase are as yet undetermined, but for convenience they are placed in a cluster. For each S haplotype, the SLFs whose products are responsible for detoxifying one or more of the five allelic variants of S-RNase tested are connected by solid arrows with their target alleles. For each S haplotype, products of one or more Other SLFs (including SLF3s of S5 and S9 haplotypes) are predicted to target products of the remaining alleles of S-RNase connected by broken arrows.[ref]
Type-3 (Primulaceae) Supergene, PrpS and PrsS
Type-3 is the gametophytic Papaveraceae-type SI, possessing the common poppy (Papaver rhoeas) stigma S (PrsS) and P. rhoeas pollen S (PrpS).[ref]
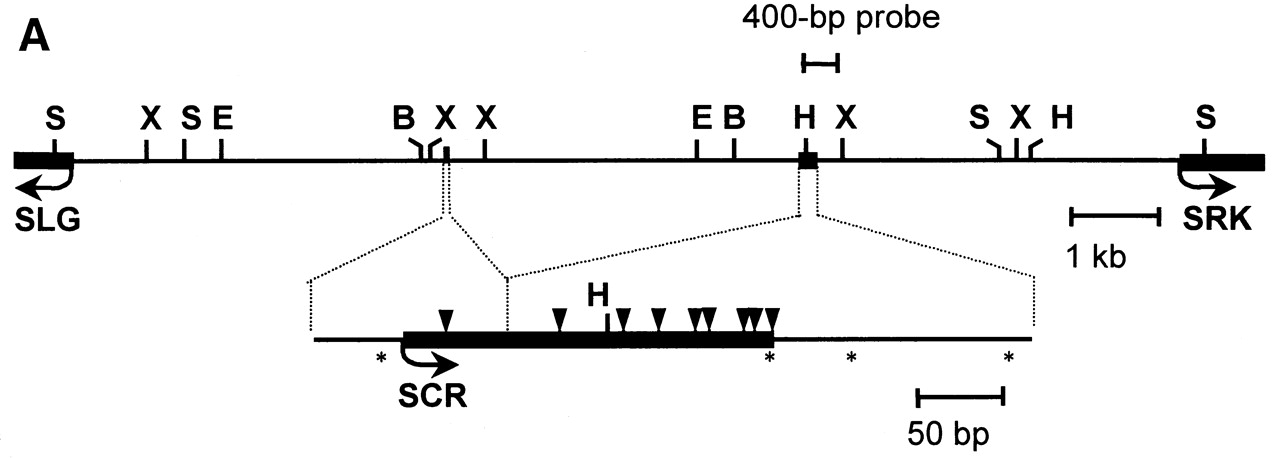
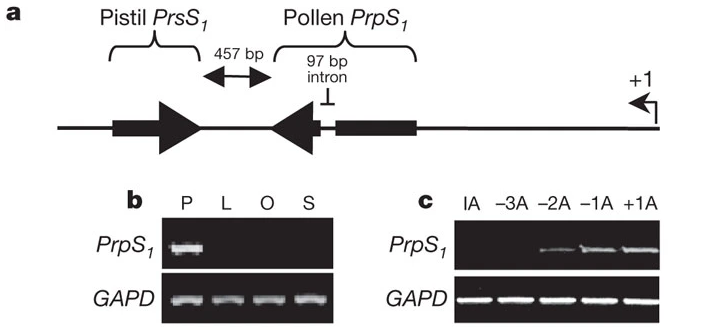
Fig. Organization of the S1 locus. Arrows indicate pistil PrsS1 and pollen PrpS1 coding sequences and their orientation.[ref]
Type-4 (Primulaceae) Supergene, CCM, CYP, GLO, KFB, and PUM
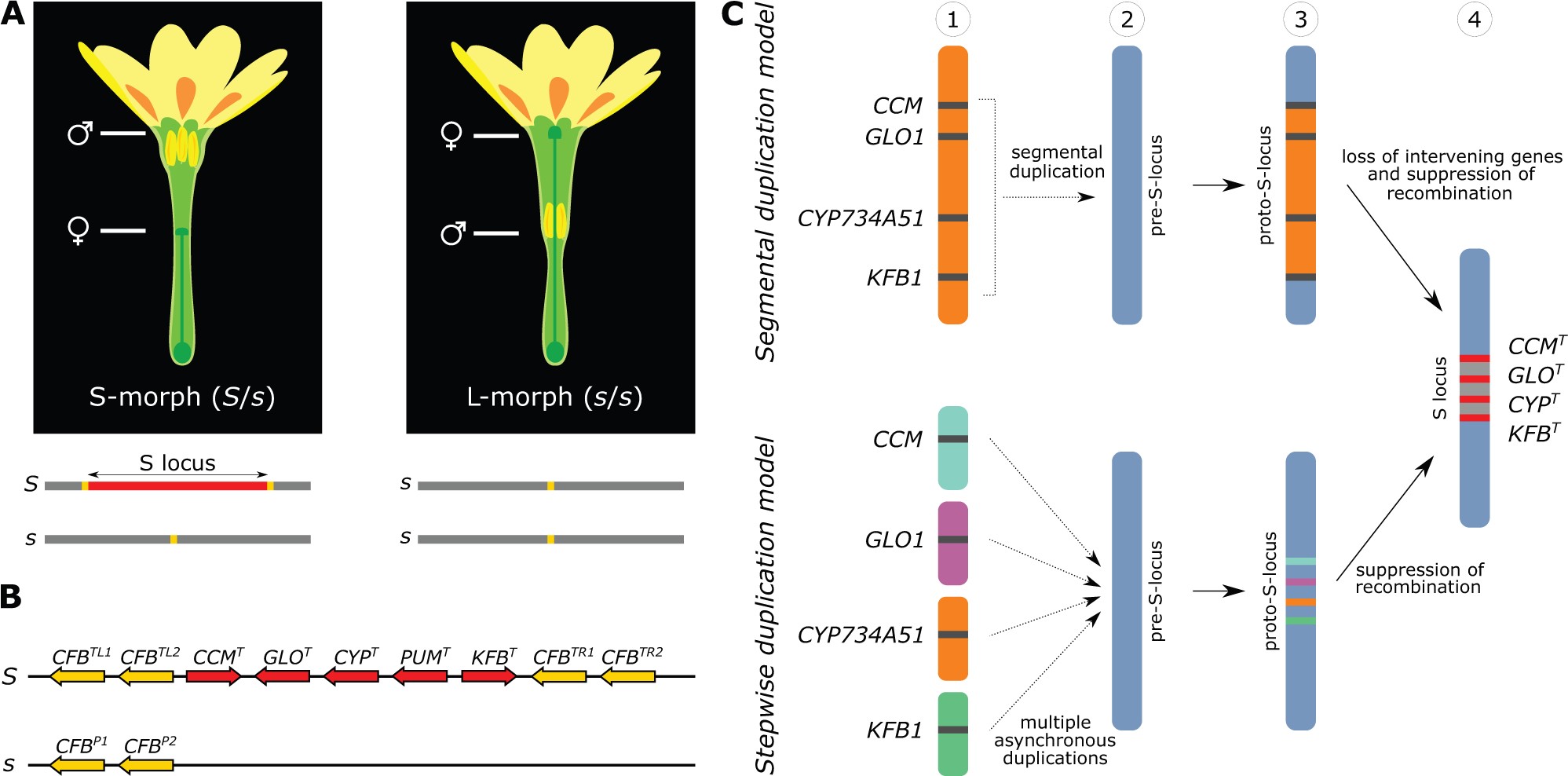
Type-4 is the sporophytic heterostyly of Primula, involving the S-locus supergene consisting of five genes encoding style length-determining cytochrome P450 (CYP450), anther position-controlling GLOBOSA (GLO), a functionally unknown Conserved Cysteine Motif (CCM), Pumilio-like RNA-binding protein (PUM) and a Kelch repeat F-Box (KFB).[ref]
Fig. Heterostyly in P. veris and models for the origin of the S locus. (A) Top: short-styled (S) and long-styled (L) morphs differ by having male (anthers) and female (stigma) sexual organs reciprocally positioned in their flowers. Bottom: The S locus (red) is hemizygous in S-morphs (S haplotype), absent in L-morphs (s haplotype); the location of CFB genes (boundry genes of S-locus) is indicated in yellow. (B) Structure of the S locus in the S-morph, with gene orientations indicated by pointed ends. Top: dominant S haplotype containing five genes (red) and two copies of CFB in each flanking region (yellow); bottom: recessive s haplotype with only two copies of CFB. Superscripts on genes stand for: T, thrum (S-morph); P, pin (L-morph); L, left; R, right; 1, gene copy 1; 2, gene copy 2. (C) Segmental duplication model (top) and stepwise duplication model (bottom) illustrated the origin and evolution of the four duplicated genes within type-4 S locus.[ref]
Type-5 (Passifloraceae) Supergene, BAHD, SPH1, and YUC6
Type-5 is the sporophytic heterostyly of Turnera, involving the S-locus supergene consisting of three genes.[ref]
Based on the well-constructed S/s haplotypes, three hemizygous genes appear to determine S-morph characteristics in T. subulata. Compared to the s-haplotype, the S-haplotype possessed three additional genes and two inversions. TsSPH1 was expressed in filaments and anthers, TsYUC6 in anthers and TsBAHD in pistils. The pistil candidate gene, TsBAHD, differs from that of Primula, but both may inactivate brassinosteroids causing short styles. TsYUC6 is involved in auxin synthesis and likely determines pollen characteristics. TsSPH1 is likely involved in filament elongation.[ref]
Fig. 1. Plants of Turnera used in this study showing species, genotypes, crosses, mutants and resources generated from them. (a) Plants used for bacterial artificial chromosome (BAC) library construction, genome and BAC sequencing, and generation of deletion mutants. (b) Genotype of self-compatible long-homostyle mutant, Drh, and photographs of L-morph, S-morph and Drh mutant flowers of T. scabra. (c) Genotype of self-incompatible short-homostyle mutant TrinSH and photographs of L-morph, S-morph and TrinSH mutant flowers of T. subulata.[ref]
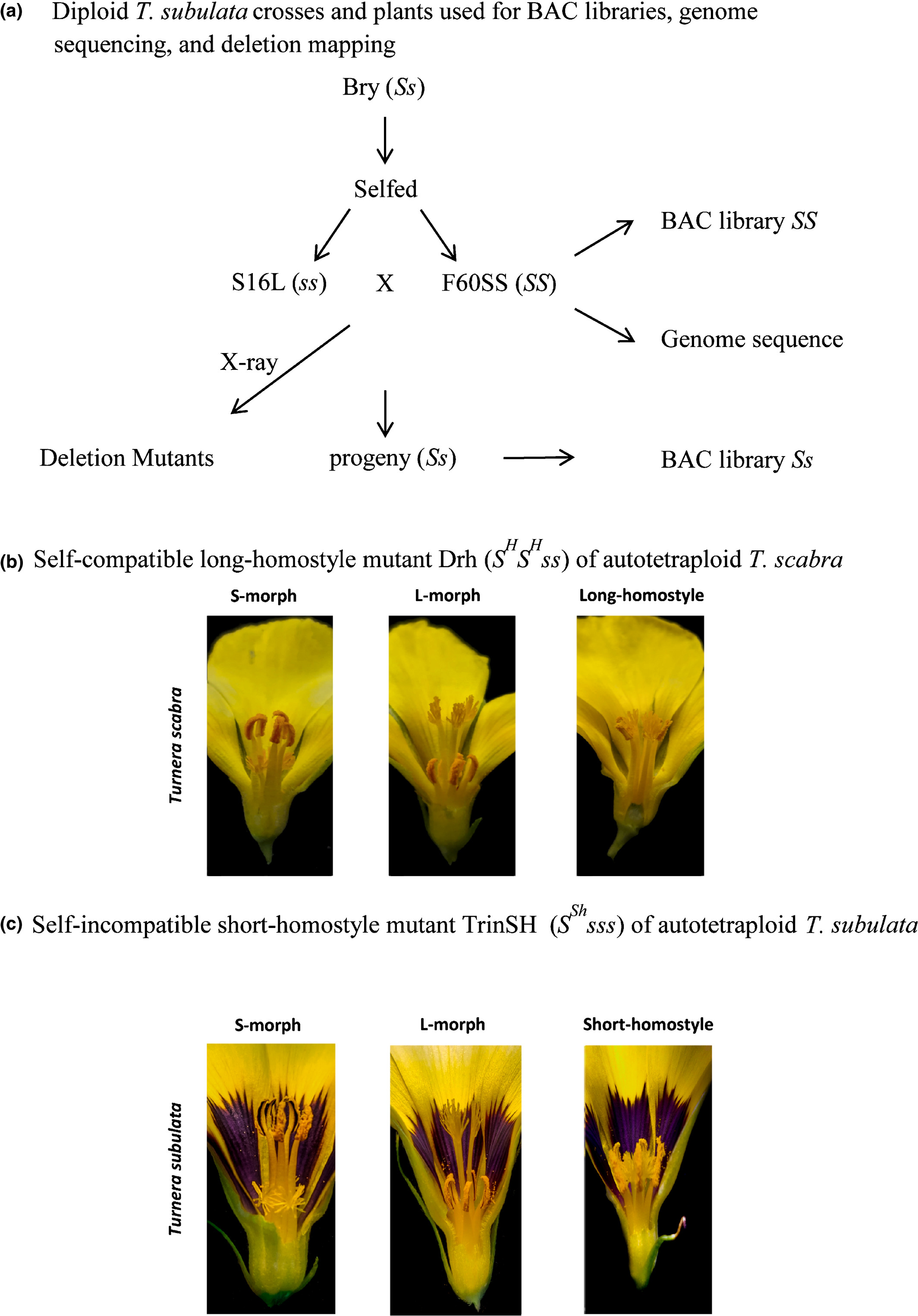
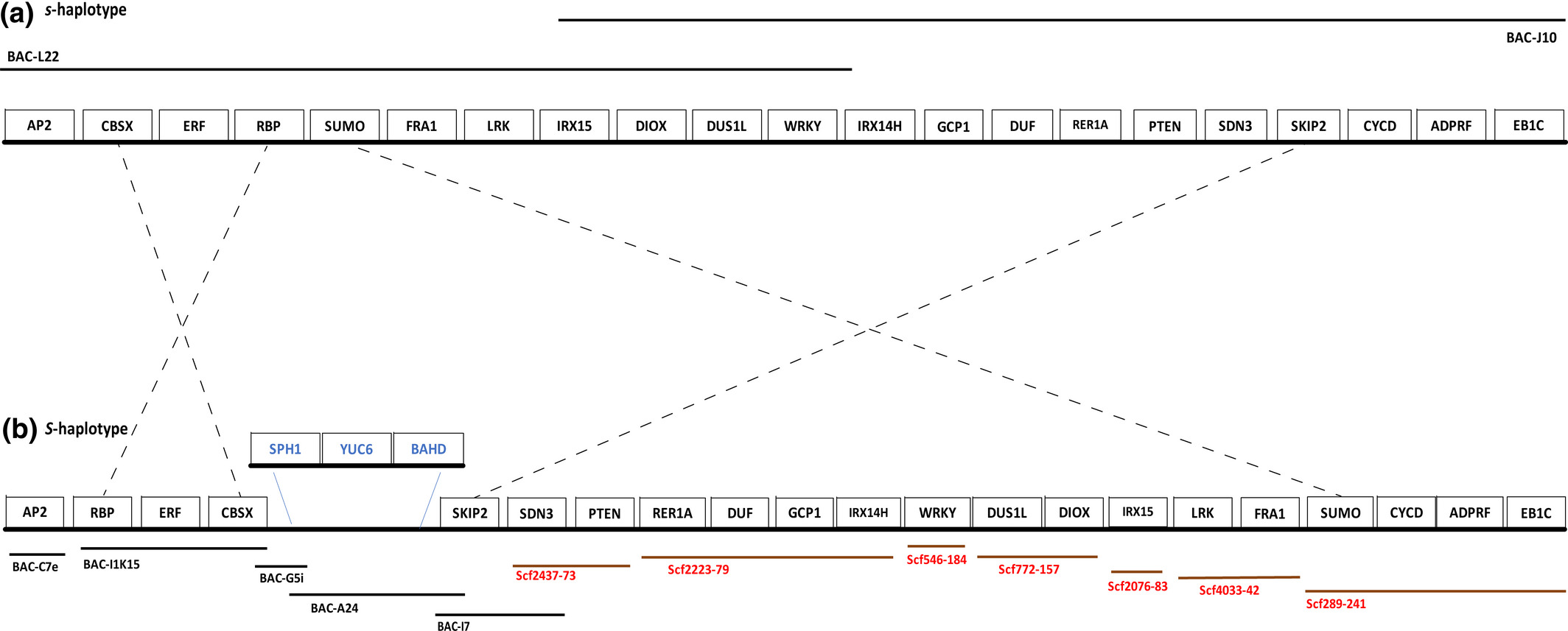
Fig. 2. Schematic showing s- and S-haplotypes of Turnera subulata including the genes they possess, the BAC clones (black lines) and/or genome scaffolds (brown lines) of which they are comprised and overlap between them. Inverted gene orders are indicated by the dashed lines. (a) The s-haplotype comprises BAC-L22 and BAC-J10. (b) The S-haplotype is comprised of five BAC clones and seven genome scaffolds. An insertion of three genes (blue font) on the S-haplotype is shown. The genome scaffold identifiers are the particular scaffold number followed by its size in kb (e.g. Scf289-241 is Scf289 which is 241 kb). Haplotypes, clones and scaffolds, are not drawn to scale.[ref]
Type-6 (Poaceae) Supergene, S-DUF247-I, S-DUF247-II, S-HPS10, Z-DUF247-I, Z-DUF247-II, and Z-HPS10
We classified the self-incompatibility of poaceae as type-6.
In the grass family (Poaceae), which contains all the cereal and major forage crops, SI has been known for half a century to be controlled gametophytically by two multiallelic and independent loci, S and Z.[ref]
Evidence showed that both the S and Z genes encodes two stamen-expressed polypeptides containing a domain of unknown function 247 (DUF247) (S- and Z-DUF247s) and a pistil-specific one possessing a signal peptide (sS/sZ).[ref]
Fig. 1. Genetic control of gametophytic self-incompatibility (GSI) by two multiple-allelic loci S and Z. When both pollen S and Z alleles are matched in the pistil, incompatibility occurs, and pollen growth is inhibited. Otherwise pollen is compatible. The degree of compatibility can be 0, 50, 75 and 100% compatible, depending on the genotypes of pollen and stigma.[ref]
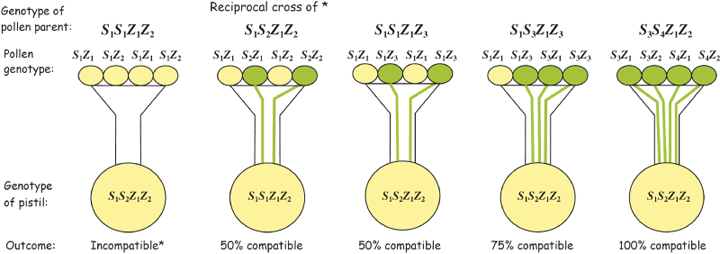
Fig. 2. Synteny maps of the Z-locus of multiple genotypes from the Poeae tribe. For the Lolium perenne L. genotype S23 Z and Lolium multiflorum Lam., both haplotypes of the diploid assemblies are given. The phylogenetic tree (left), representing the different species, was drawn according to the NCBI taxonomy database. The gene- and marker-less regions are represented as shaded breaks, and a white space indicates an assembly gap. The genes present at the Z-locus are represented with directed arrows, and self-incompatibility candidate genes are colored in teal and blue. The gene orientation is shown with the pointy side representing the 3′ end. The markers used for the fine-mapping are represented by black bars, and the number of recombinants for each marker is indicated between brackets for L. perenne Kyuss and F1-30. The synteny between genes is illustrated by lines, and in case of orientation change, a small circular arrow is used. In addition, the compatibility phenotype of the genotype is indicated on the right: self-incompatible (SI) or self-compatible (SC).[ref]
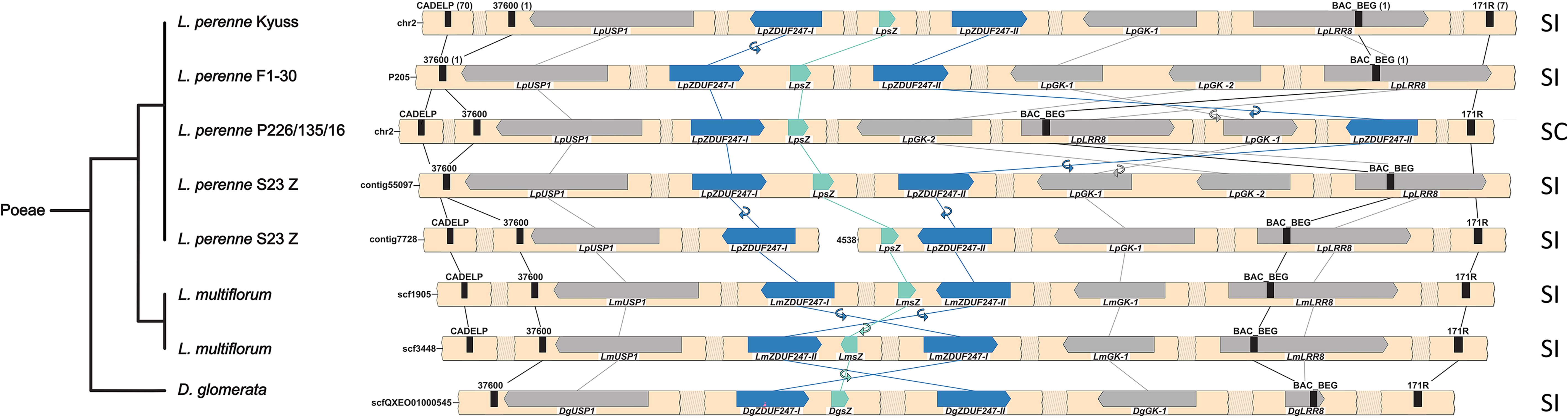
Fig. 3. Synteny maps of the S-locus of multiple genotypes from the Poeae tribe. For the Lolium perenne L. genotype S23 Z and Lolium multiflorum Lam., both haplotypes of the diploid assemblies are given. The phylogenetic tree (left), representing the different species, was drawn according to the NCBI taxonomy database. The gene- and marker-less regions are represented as shaded breaks, and a white space indicates an assembly gap. The genes present at the S-locus are represented with directed arrows, and the self-incompatibility candidate genes are colored in teal and blue. The gene orientation is shown with the pointy side representing the 3′ end. The markers used for the fine-mapping are represented by black bars, and the number of recombinants for each marker is indicated between brackets for L. perenne Kyuss and P226/135/16. The synteny between genes is illustrated by lines, and in case of orientation change, a small circular arrow is used. In addition, the compatibility phenotype of the genotype is indicated on the right: self-incompatible (SI) or self-compatible (SC).[ref]
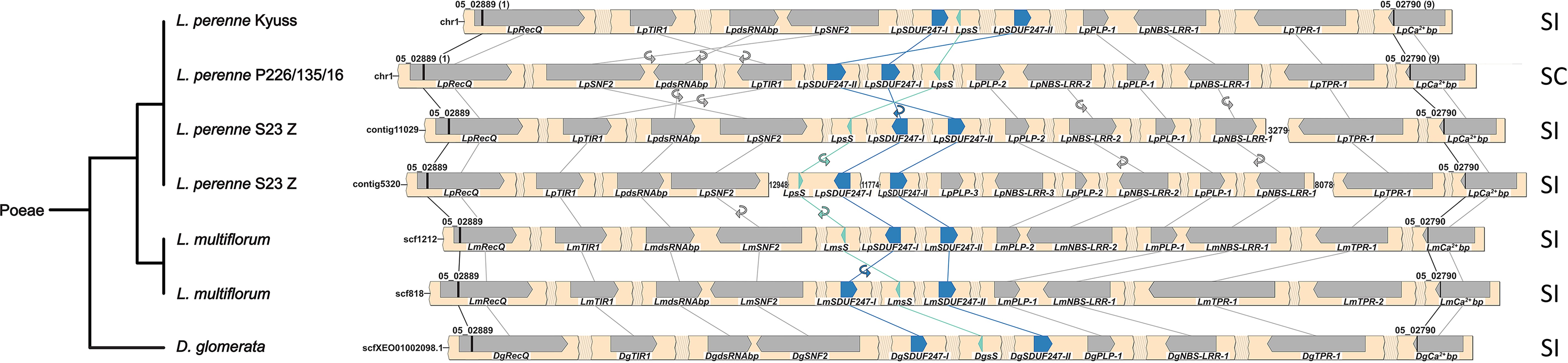
Type-7 (Linaceae) Supergene, TSS1 and WDR-44
We classified the self-incompatibility of Linaceae as type-7.
The S-locus of Linum tenue was identified using population genomic data. The results show that hemizygosity and thrum-specific expression of S-linked genes, including a pistil-expressed candidate gene for style length, are major features of the Linum S-locus. Two distyly candidate genes (LtTSS1 and LtWDR-44) are only present in the dominant S allele.[ref]
Fig. 1. Distyly in Linum tenue. A) Thrum individuals (also termed S-morph, to the left) are short styled, whereas pin individuals are long styled (L-morph, to the right). (B) Reciprocal location of male (anther) and female (stigma) reproductive organs in thrum (left) and pin (right) flowers. Petals and sepals have been removed to better visualize reproductive structures, and the scale is indicated by the scale bar (5 mm). The expected S-locus genotype is indicated below each morph.[ref]

Fig. 2. Identification of the distyly S-locus based on population genomic analyses. (A) Differences in coverage between thrum (n = 26) and pin (n = 25) samples indicates the existence of an ∼260-kb hemizygous region in LG10 (shaded regions indicate 95% CI), as supported by the existence of six 50 kb adjacent windows that are absent in pin samples. (B) Genetic differentiation (FST) between morphs in 5-kb windows flanking the ∼260-kb hemizygous region, using samples from population SMT (thrum = 13 and pin = 13) (see estimates for population CL; Figure S4). Windows with statistically significant estimates are highlighted in red (p < 0.01, after 1,000 replicates of permutation test per window, followed by FDR correction with the Benjamini-Hochberg method). (C) Differences in mean π between thrum and pin individuals, also in 5 kb windows flanking the ∼260 kb hemizygous region for population SMT and with statistically significant estimates highlighted in red. [ref]
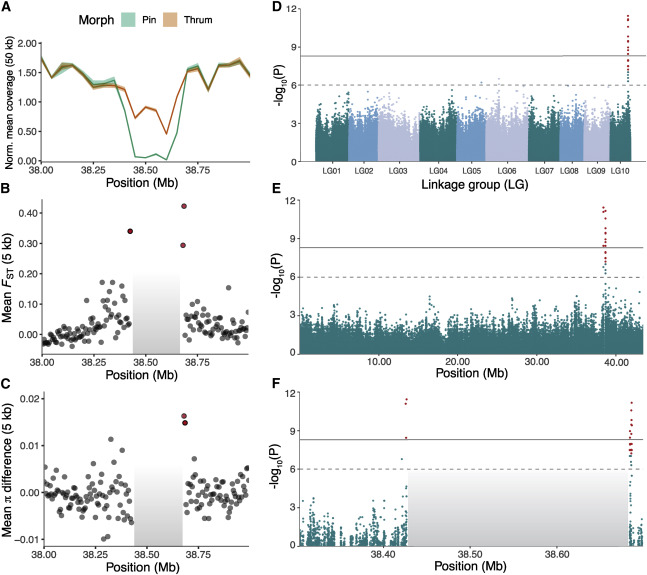
Fig. 3. Haplotype structure and patterns of molecular evolution at the S-locus. (A) Schematic representation of the dominant S and recessive s alleles of the distyly S-locus, indicating the location of the two distyly candidate genes LtTSS1 and LtWDR-44, only present in the dominant S allele.[ref]
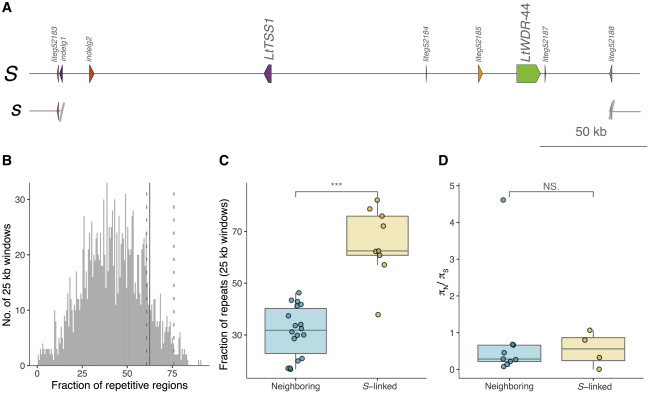
Type-8 (Oleaceae) Supergene, GA2ox
We classified the self-incompatibility of Oleaceae as type-8.
The S-locus region of Phillyrea angustifolia proved to have a segregating 543-kb indel unique to one specificity, suggesting a hemizygous region, as observed in all distylous systems so far studied at the genomic level. Only one of the predicted genes (GA2ox-S) in this indel region is found in the olive tree. The presence/absence polymorphism of the GA2ox-S gene is stably associated with SI groups across Oleaceae.[ref]
Fig. 1. Chromosomes controlling the [Hb] specificity carry a 543-kb indel. The candidate 5-Mb interval contains 65 predicted protein-coding genes, and a 543-kb indel (box delimited by dotted line) spanning over six genes contains a large number of [Hb]-specific sequences. Haplotype 1 of the (SS) individual was aligned on haplotype 2 (non-inverted) of the (ss) individual. Gray areas delimited by thin black lines connect aligned portions of the two chromosomes. Predicted protein-coding genes are represented by black vertical lines along each of the two chromosomes. The s- and S-specific 300-bp sequences are represented by blue and red vertical lines, respectively.[ref]
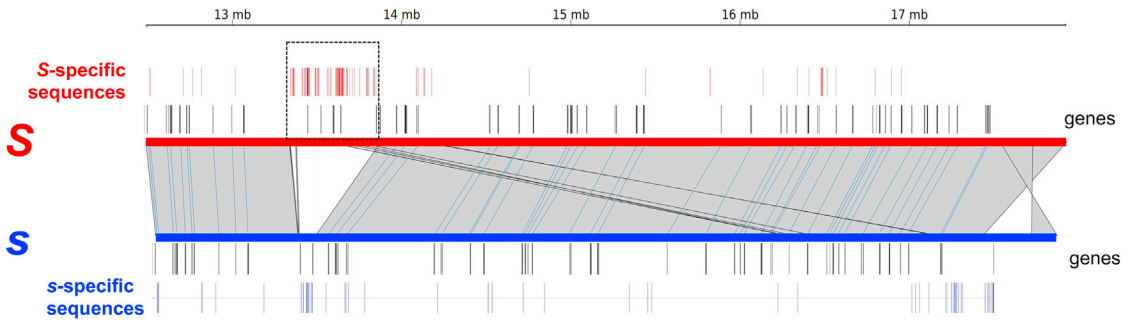
Fig. 2. The indel in P. angustifolia contains six predicted protein-coding genes. Comparison of the indel sequence between P. angustifolia and O. europaea (var. Arbequina) reveals that GA2ox-S is the only conserved gene in the indel, with high divergence of intergenic sequences. Short reads mapping of O. europaea accessions (at the bottom of the figure, y axis indicates read mapping depth up to 100X) identifies a segregating 756-kb indel in the Arbequina chromosome 18 assembly. Triangles indicate annotated genes that putatively correspond to transposable elements. Crosses indicate genes with no sequence similarity (by blast) with anything in the chromosomal fragment of the other species. Solid lines (black and red) indicate orthologous genes. Interrupted lines indicate genes in one species with strong sequence similarity but no gene annotation in the chromosomal fragment of the other species.[ref]
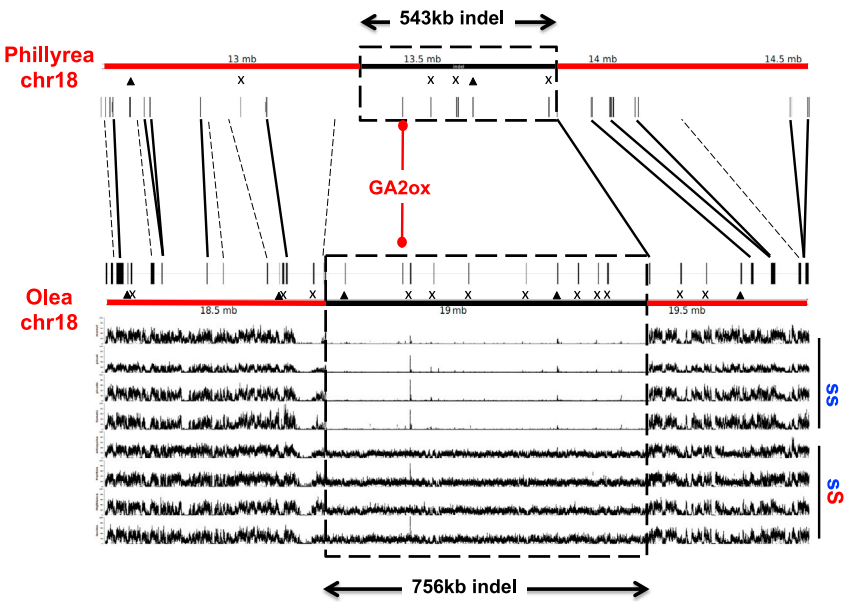
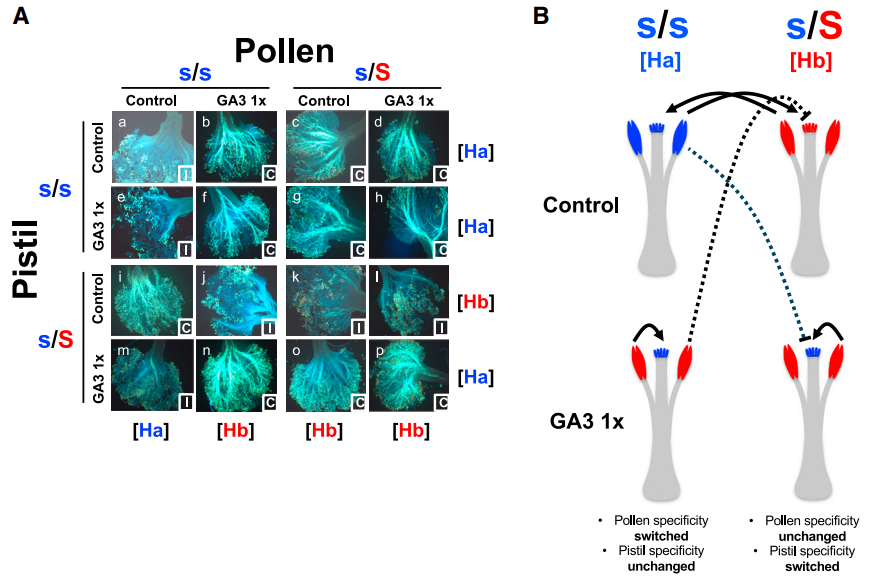
Fig. 3. Treatment with gibberellin modifies the male and female SI responses of the two specificities in different manners. (A) Fluorescent microscopic images of controlled pollination assays in Ligustrum vulgare. Compatible and incompatible reactions (indicated in the bottom right corner of each image) are identified based on the presence of germinated pollen tubes reaching the style. (B) Graphical summary of the effect of the GA supplementation assay, illustrating that pistils of the [Ha] specificity and pollen of the [Hb] specificity were GA insensitive, while, conversely, pollen of the [Ha] specificity and pistils of the [Hb] specificity switched specificity upon GA3 treatment.[ref]