User Manual
Database Overview
1. Overview of PSI Atlas
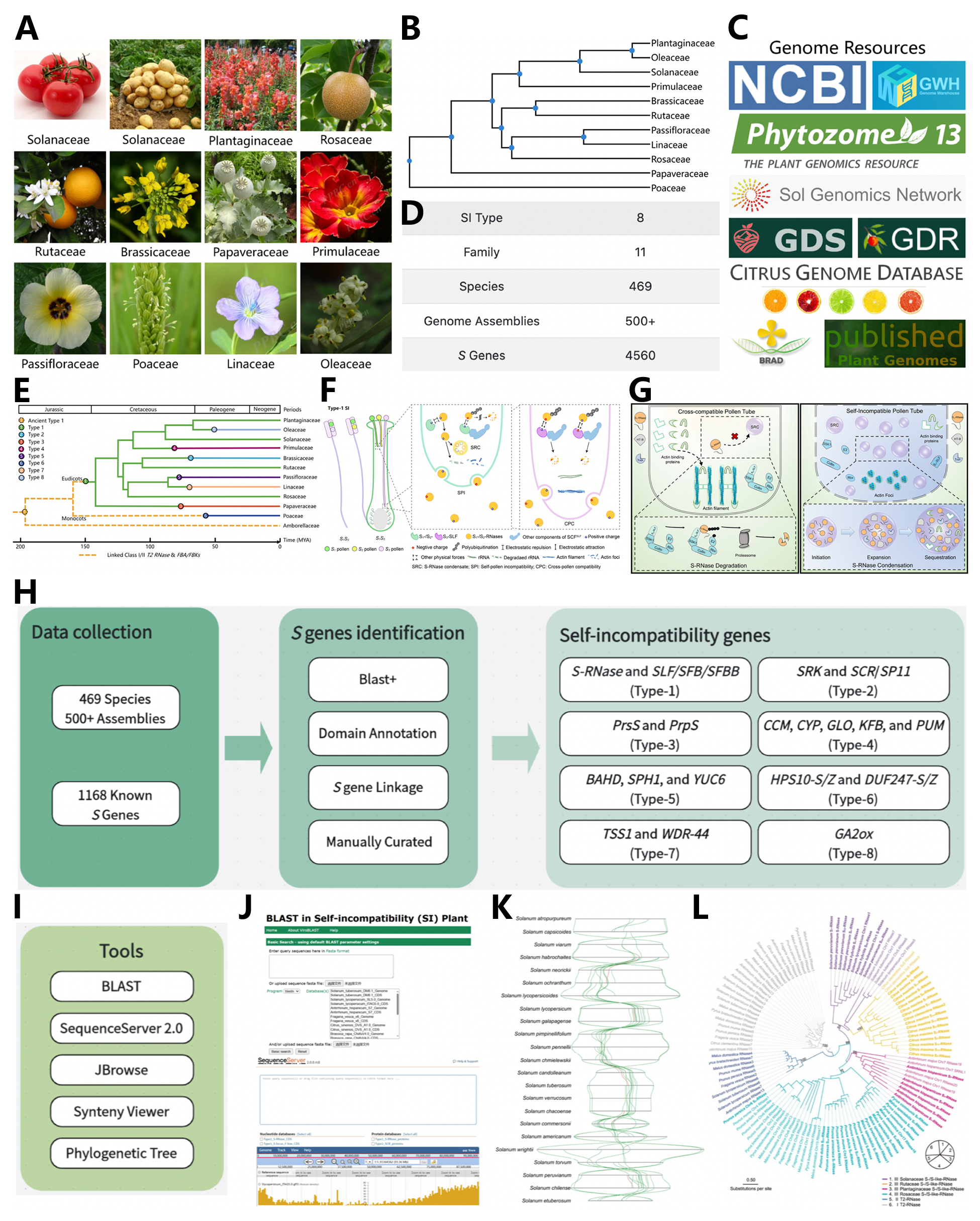
A. Representative species of eight self-incompatibility (SI) types. B. The phylogenetic relationships of all existing families in PSIA. C. Genome resources of SI species. D. Overview of SI data in the latest release. E. The origin and evolution of the eight SI types. F. Diagram illustrating the molecular mechanism of type-1 SI with those of all eight types shown in PSIA. G. The reported working model of SI found in the representative species Petunia hybrida. H. The pipeline of S gene identification. I. Useful tools of PSIA. J. BLAST and JBrowse function of PSIA. K. The output of the synteny analysis between the Solanum S-loci is shown as an example. L. The output of the phylogenetic analysis for S-RNases from the exemplar families such as Plantaginaceae, Solanaceae, Rosaceae and Rutaceae.
2. Browse the 'home' page for quickly start
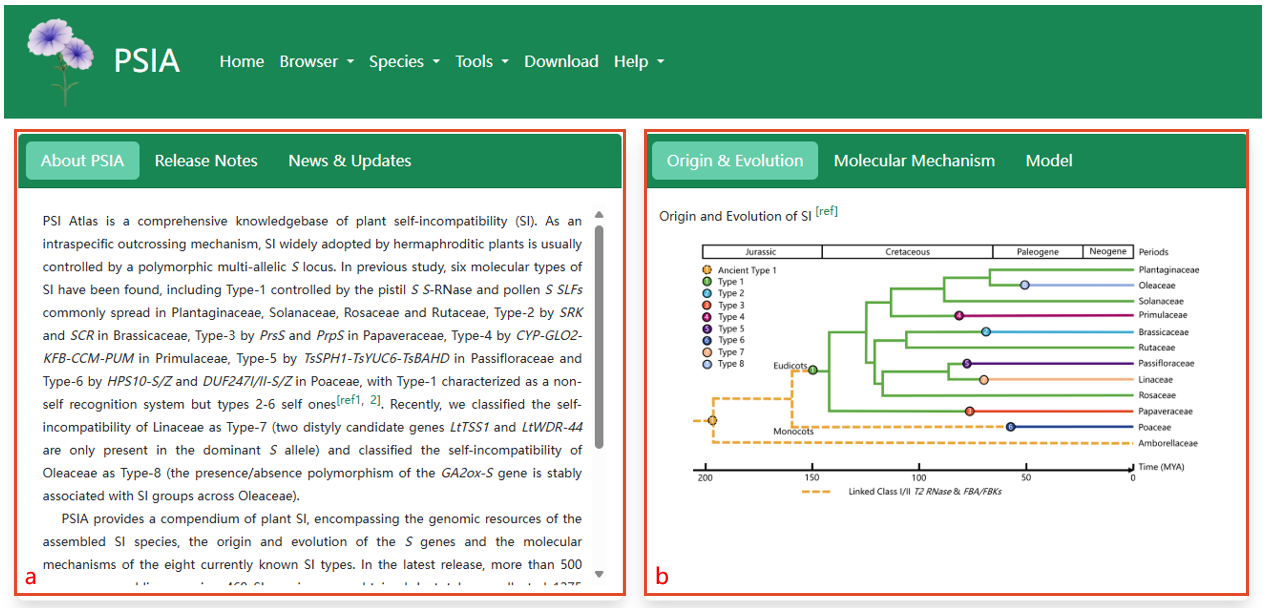
a. Brief information of PSIA, release note of genome assemblies, news and updates of PSIA. b. The origin and evolution of the eight SI types, eight diagrams of different molecular mechanism types, two reported working models of SI found in the representative species Petunia hybrida.
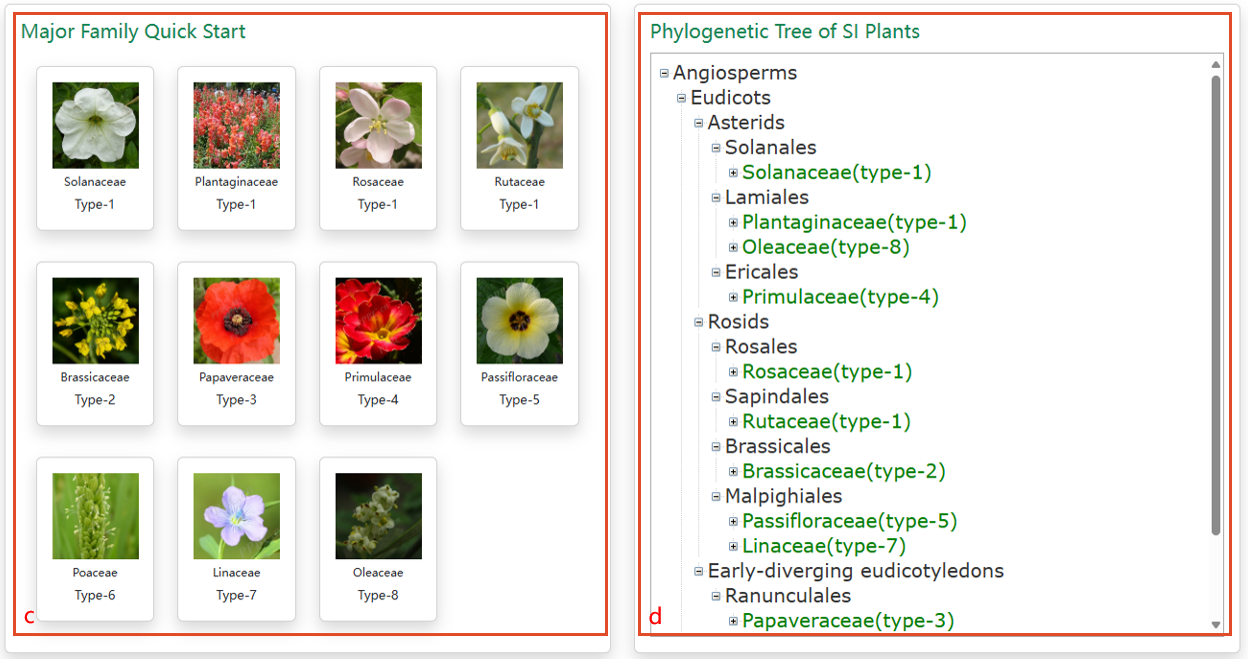
c. Quick start links of 11 SI families. d. Phylogenetic positions of SI plants with genomes sequenced and published.
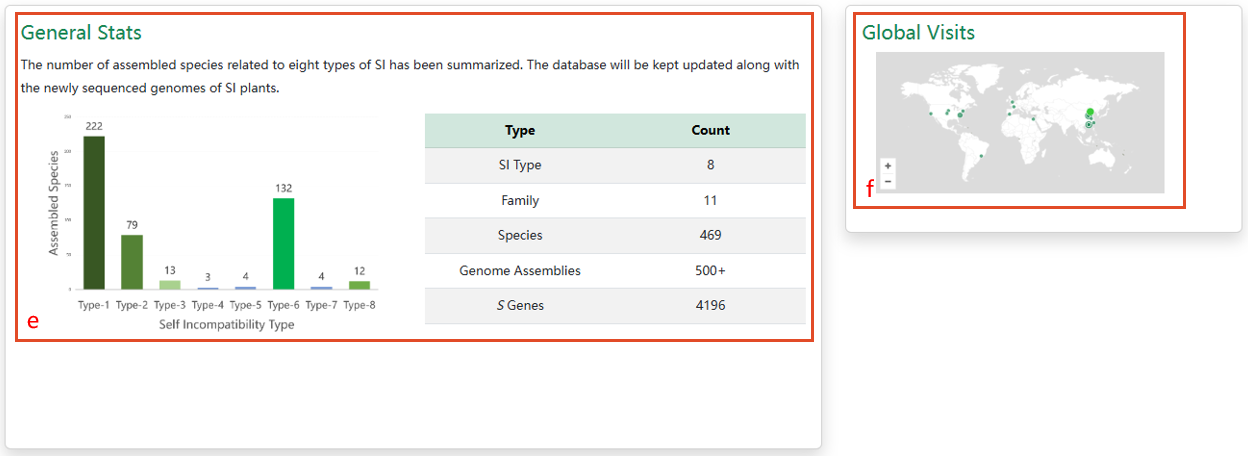
e. The latest statistics of PSIA. f. Global visits map.
Data Curation
1. Genome assemblies resources
PSIA has the most comprehensive genome assembly information of SI plants, with all available genome assemblies of SI species collected from NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/), NGDC-GWH (Genome Warehouse, https://ngdc.cncb.ac.cn/gwh), Published Plant Genomes (https://www.plabipd.de/plant_genomes_pa.ep), Phytozome v13 (https://phytozomenext.jgi.doe.gov), Sol Genomics Network (https://solgenomics.net), Genome Database for Rosaceae (https://www.rosaceae.org), Genome Database for Strawberry (http://eplant.njau.edu.cn/strawberry/), Citrus Genome Database (https://www.citrusgenomedb.org), Brassicaceae Database (http://www.brassicadb.cn/#/), and other links (such as data availability links in papers, figshare, and google drive). In the current release, more than 500 genome assemblies covering 469 SI species were obtained. Besides, our database will be regularly updated to incorporate the latest sequenced genomes of SI plants.
2. Supergene identification
The S-locus information of the assembled SI species has been fully investigated in PSIA based on the known molecular types. We started by downloading all known S genes from the NCBI Nucleotide and Protein Database and totally collected 1,275 nucleotide and 1,130 protein accessions of S genes from public resources. Next, we used blast+ to find S genes in all of the collected genome assemblies of SI plants, and manually curated the coding regions of each gene to ensure accurate splice sites. For S determinants containing conserved secondary domains, such as the F-box and FBA/FBK of SLF/SFB/SFBB and the Ribonuclease T2 family domain of S-RNase, we further searched for them to identify the candidate type-1 S. For type-2 to type-8 SI, S genes were identified according to the blast results and linkage distance as previously described. In total, 3,285 S genes have been identified and curated manually in PSIA.
Knowledge of SI origin, evolution, molecular mechanisms and working models
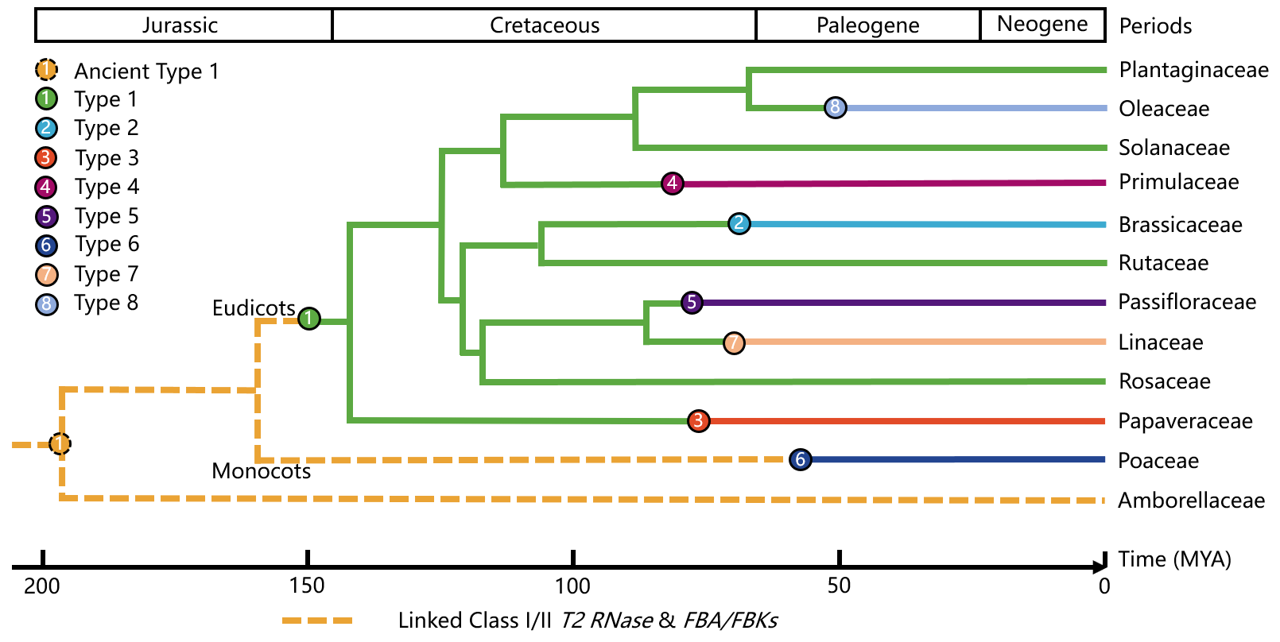
1. Origin and evolution of SI
The phylogenetic tree of species at the family level illustrating the origin of the eight SI types. The evolutionary tree is generated by TimeTree. The geological time scale is shown at the top, while the bottom numerical axis represents the evolutionary timeline. The circles of different colors and their corresponding lines represent the eight types of SI. The orange dashed line indicates the ancient type-1 S structure.
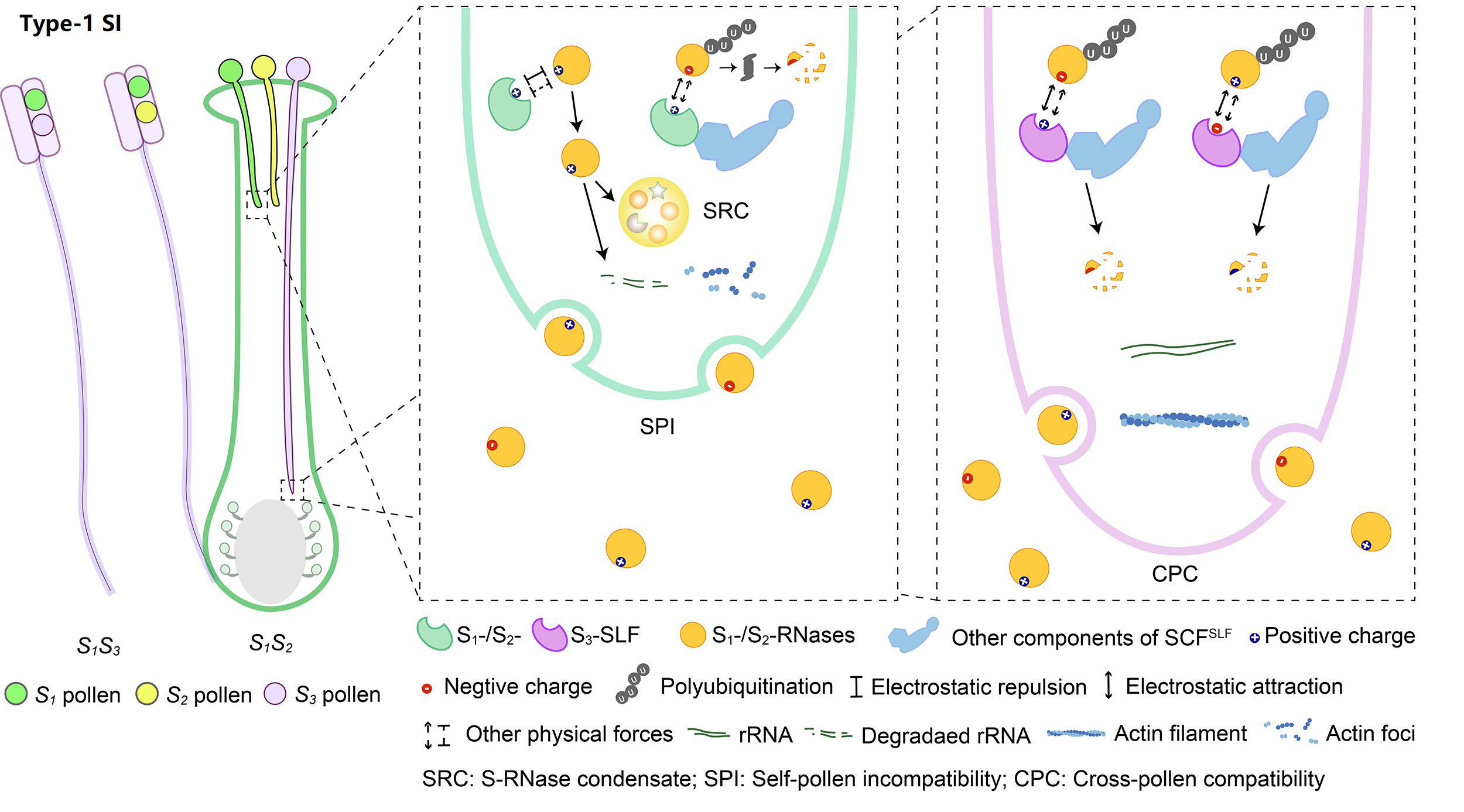
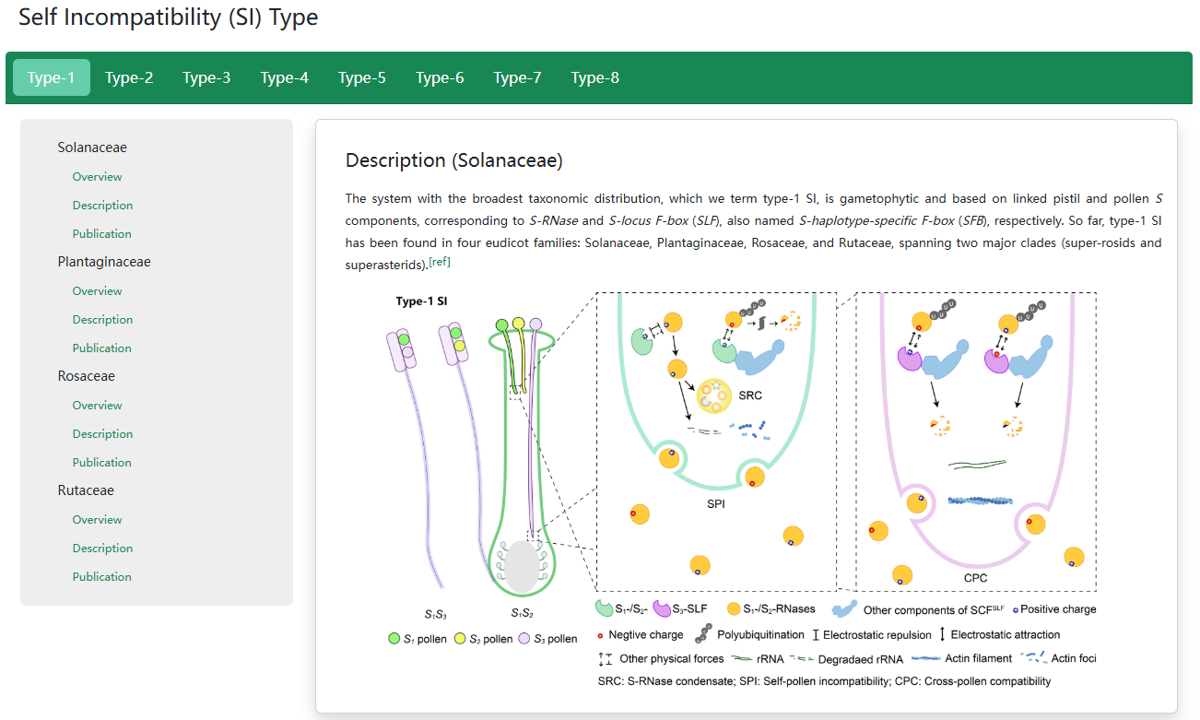
2. Molecular mechanism of Type-1 SI
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.
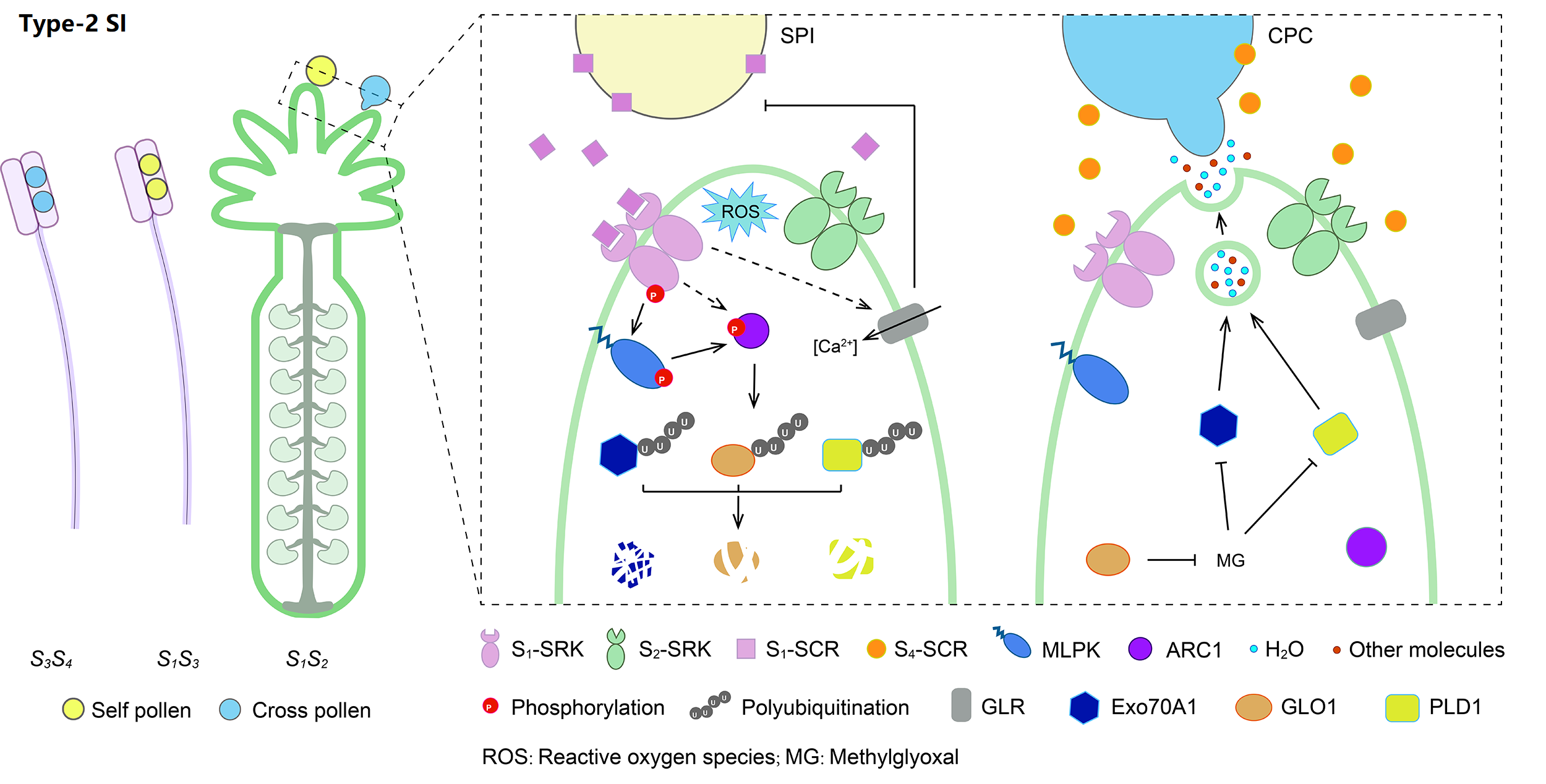
3. Molecular mechanism of Type-2 SI
Type-2 SI is the sporophytic Brassicaceae-type SI, controlled by a male S-locus cysteine-rich (SCR) protein/S-locus protein 11 and a female S-locus receptor kinase (SRK). Pollen S1-SCR can specifically recognize its cognate S1-SRK after pollination, triggering several signaling cascades mediated by phosphorylation and ubiquitin-proteasome system, thus leading to self-pollen rejection.
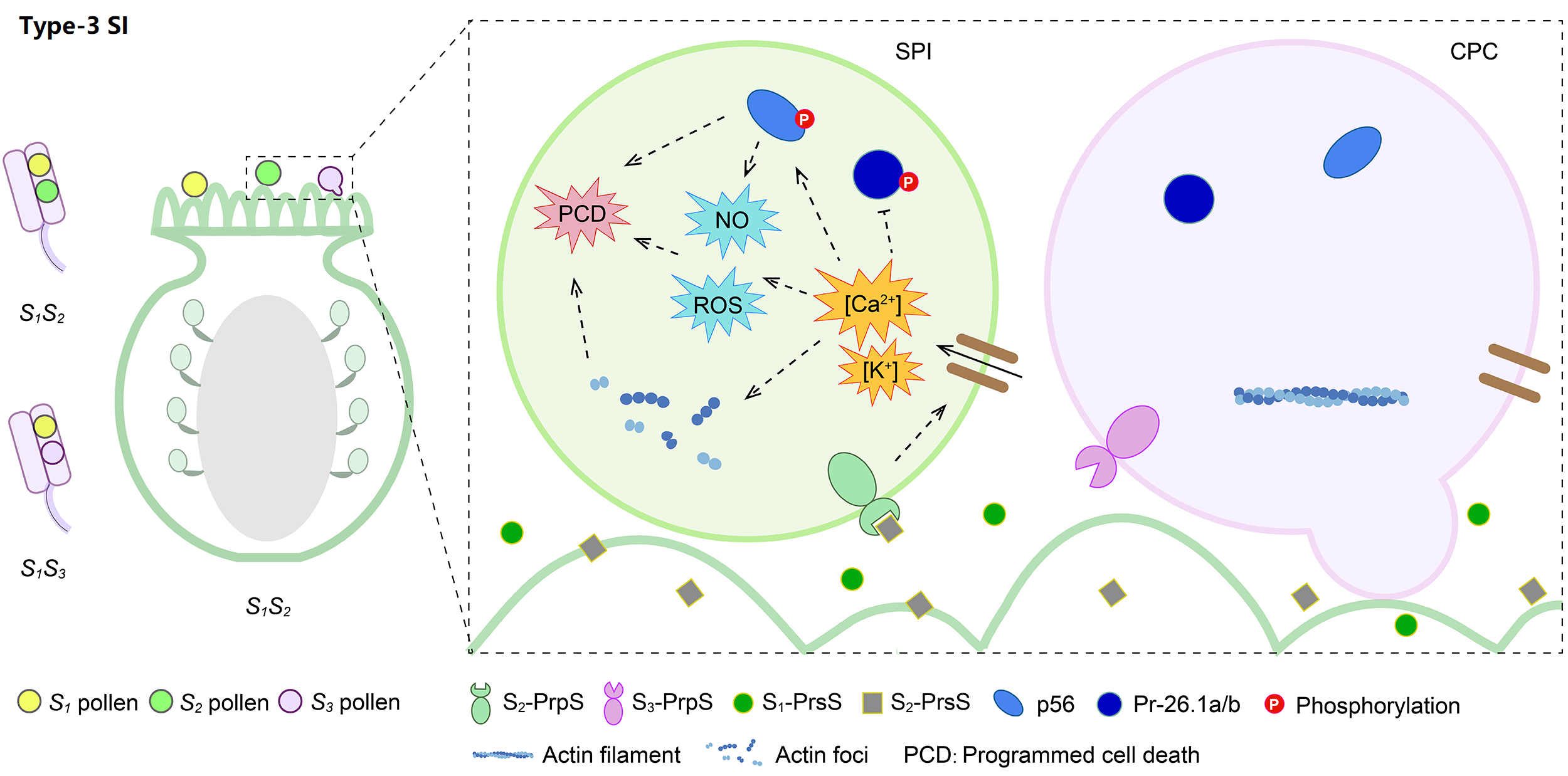
4. Molecular mechanism of Type-3 SI
Type-3 is the gametophytic Papaveraceae-type SI, possessing the common poppy (Papaver rhoeas) stigma S (PrsS) and P. rhoeas pollen S (PrpS). Prs S2 secreted by S1S2 papilla cells can specifically bind to pollen membrane-localized Prp S2, stimulating Ca2+ influx, ROS accumulation, actin depolymerization, and PCD of self-pollen.
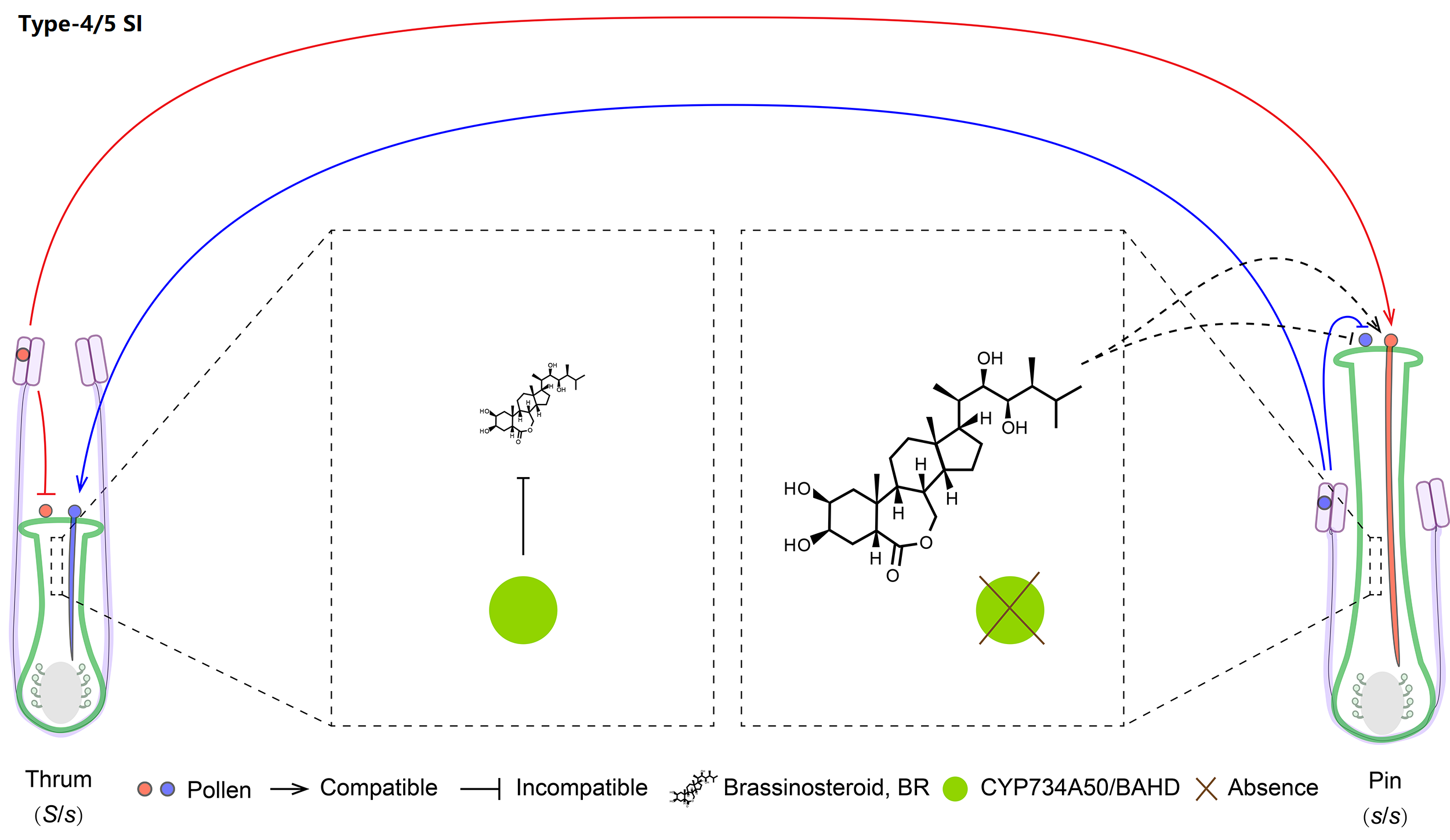
5. Molecular mechanism of Type-4/5 SI
Type-4 and -5 are the sporophytic heterostyly SI of Primulaceae and Turneraceae, controlled by hemizygous S-loci mainly encoding CYP (Cytochrome P450) and TsBAHD, respectively. Athough both of them can inactivate pistil BR, they are absent in the long styles, leading to nonself pollen acceptance and self pollen rejection.
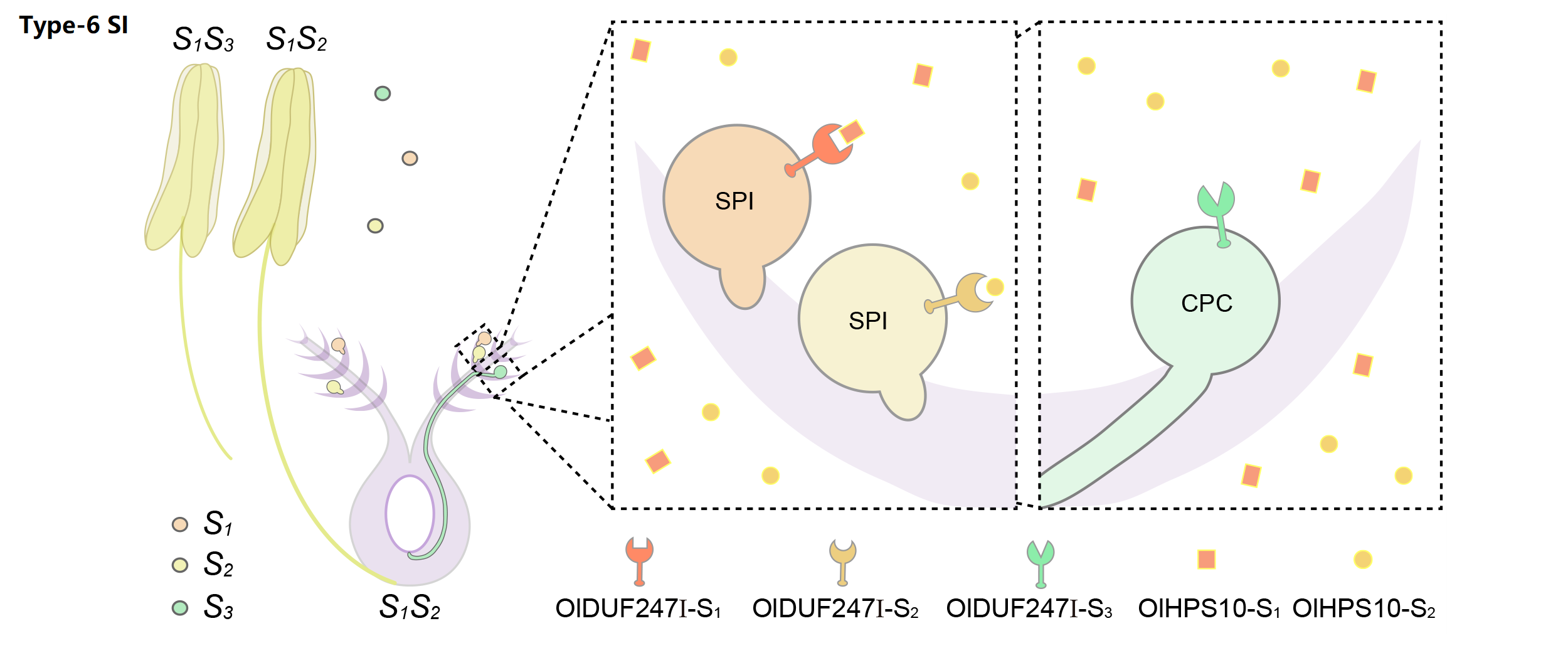
6. Molecular mechanism of Type-6 SI
We classified poaceae SI as type-6 gametophytically controlled by two multiallelic and independent loci, S and Z. Taking S as an example, stamen-expressed transmembrane polypeptides containing a domain of unknown function 247 (DUF247) can specifically recognize the pistil-specific peptide HPS10, resulting in the rejection of the self pollen.
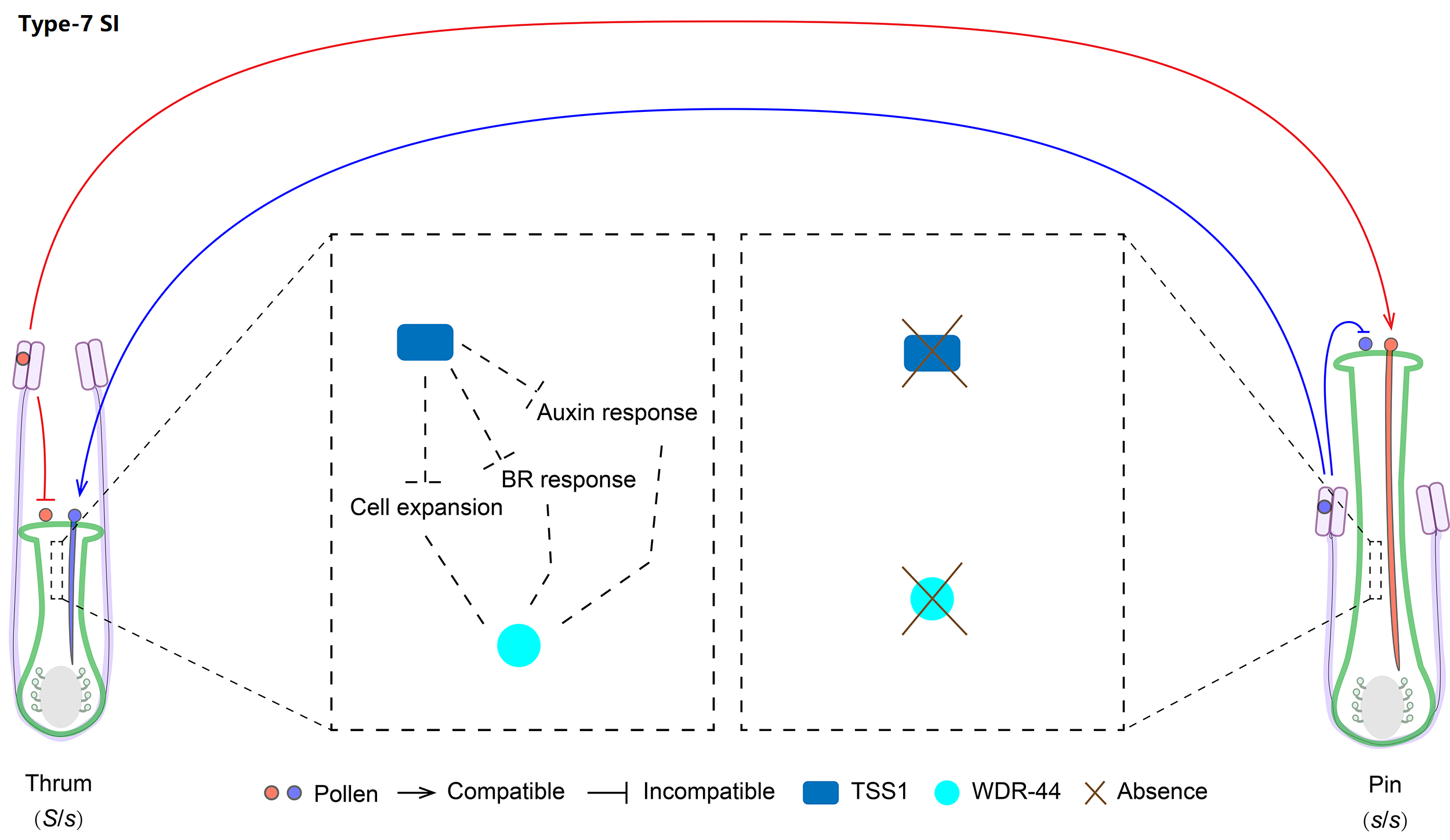
7. Molecular mechanism of Type-7 SI
We classified Linaceae heterostyly SI as type-7 with two distyly candidate genes (LtTSS1 and LtWDR-44) only expressed by the dominant S allele, which can further regulate cell expression, BR and auxin responses of the short styles.
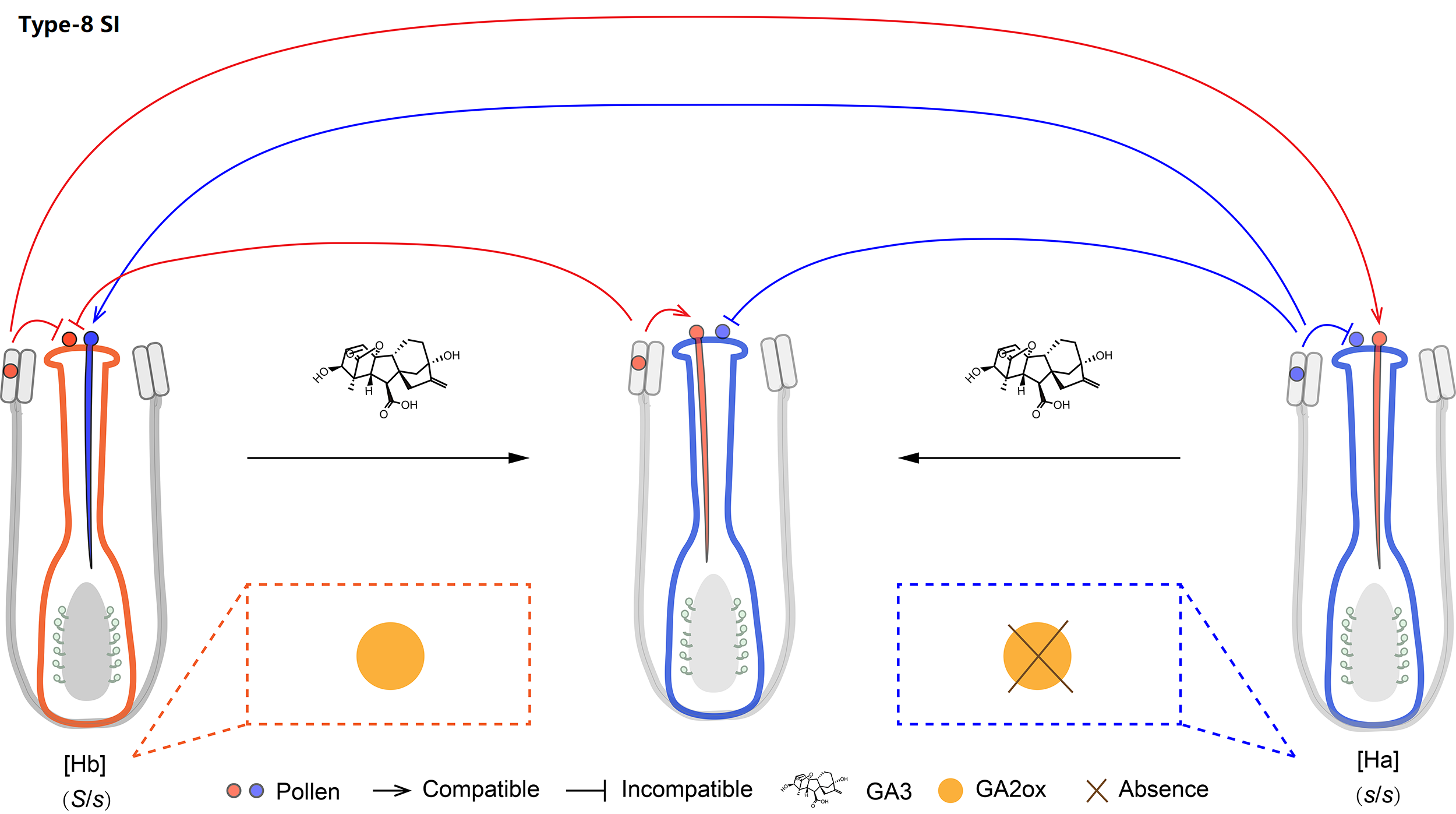
8. Molecular mechanism of Type-8 SI
We classified Oleaceae SI as type-8. The presence/absence polymorphism of the GA2ox-S gene is stably associated with SI groups across Oleaceae. GA3 treatment can switch the female specificity of [Hb] individuals and the male specificity of [Ha] ones.
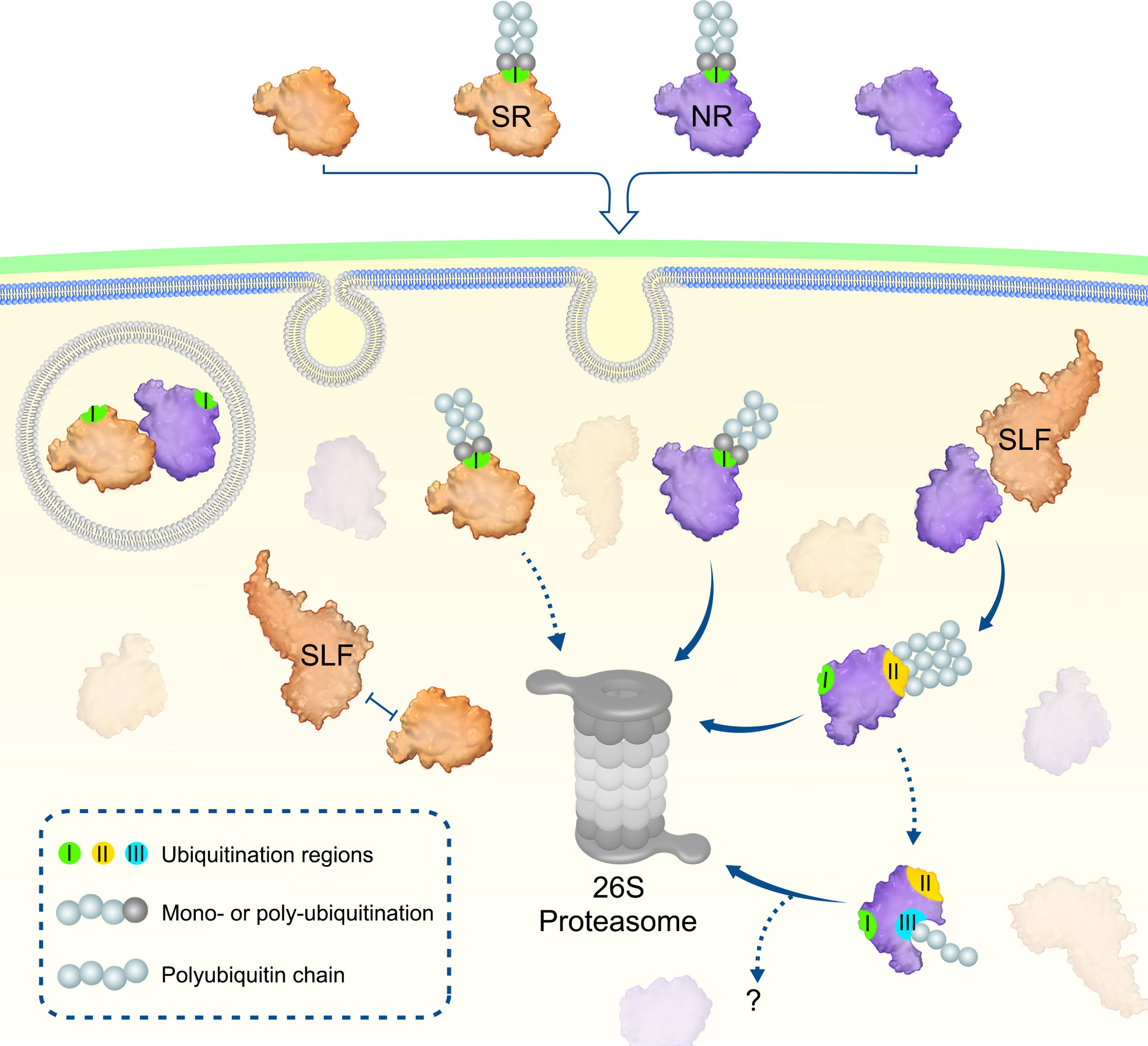
9. Proposed model for a stepwise ubiquitination and degradation mechanism of Petunia hybrida S-RNases[ref]
Proposed model for a stepwise ubiquitination and degradation mechanism of Petunia hybrida S-RNases. Self and nonself S-RNases (SR and NR) and their region (R) I ubiquitinated forms can enter the cytoplasm through the pollen-tube membrane. The R I ubiquitinated S-RNases could be degraded by the 26S proteasome, whereas their unubiquitinated forms are identified by SLFs. SR and its cognate SLF repel each other and results in an inability of SCFSLF to ubiquitinate SR. By contrast, NR is attracted by nonself SLF for R II ubiquitination by SCFSLF and its subsequent degradation by the 26S proteasome. Subsequently, internal R III could be further exposed for ubiquitination, leading to degradation of NR by the 26S proteasome or other unknown pathways. In addition, S-RNase compartmentalisation could occur in the vacuole and contribute to its sequestration. I, II, and III: three ubiquitination regions in S-RNases[ref].
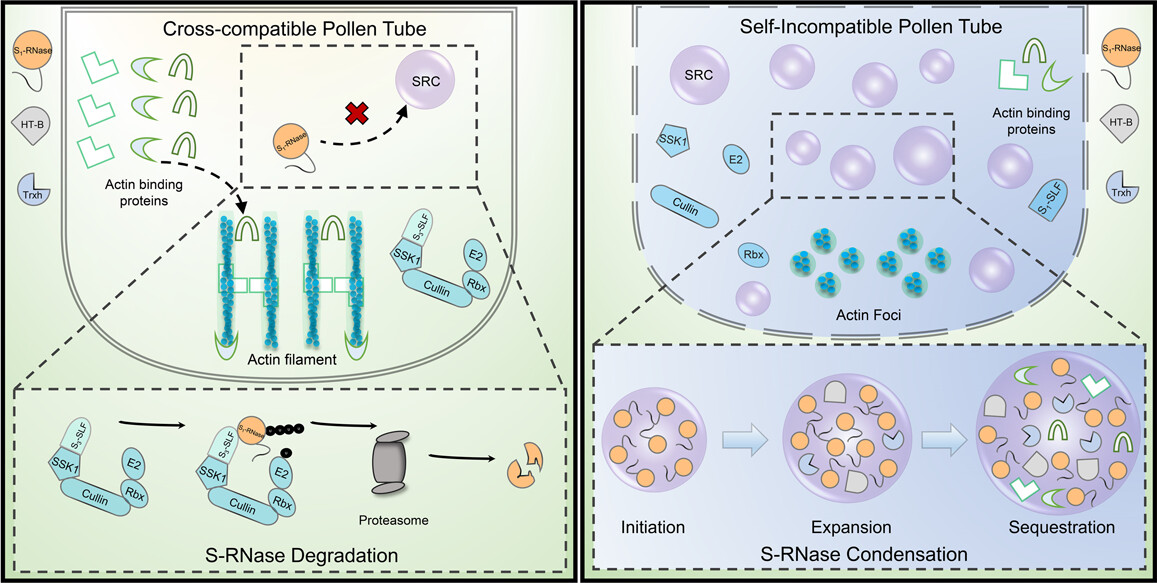
10. Model of self-incompatibility in Petunia hybrida[ref]
In the cross-compatible pollen tube, S1-RNases, HT-B, and thioredoxin h (Trxh) enter into the non-self-pollen tube. S3-SLFs (S-locus F-boxes) form Skip Cullin F-box (SCF) complexes with SSK1, Cullin1, and Rbx to degrade S1-RNase by the ubiquitin–proteasome system (UPS). Meanwhile, actin-binding proteins have normal abundance and can bind to the cytoskeleton to maintain the order of the cytoskeleton, resulting in non-self-pollen tube growth. In the self-incompatible pollen tube, S1-RNases, HT-B, and Trxh enter into the self-pollen tube. Self S1-RNase cannot be recognized and degraded by the SCF complex, thus remaining functional and forming condensates (Initiation). Further, S1-RNase together with HT-B and Trxh forms robust S-RNase condensates (SRCs) (Expansion), which further recruit actin-binding proteins (Sequestration). The sequestration of those factors by SRCs causes the cytoskeleton to be disordered due to the lack of actin-binding proteins, generating actin foci and inhibiting self-pollen tube growth. The dashed boxes at the bottom indicate the S-RNase degradation and condensation processes, respectively. Non-self- and self-pollen tubes are generally in oxidation (yellow) and reduced (blue) states, respectively[ref].
SI Browser
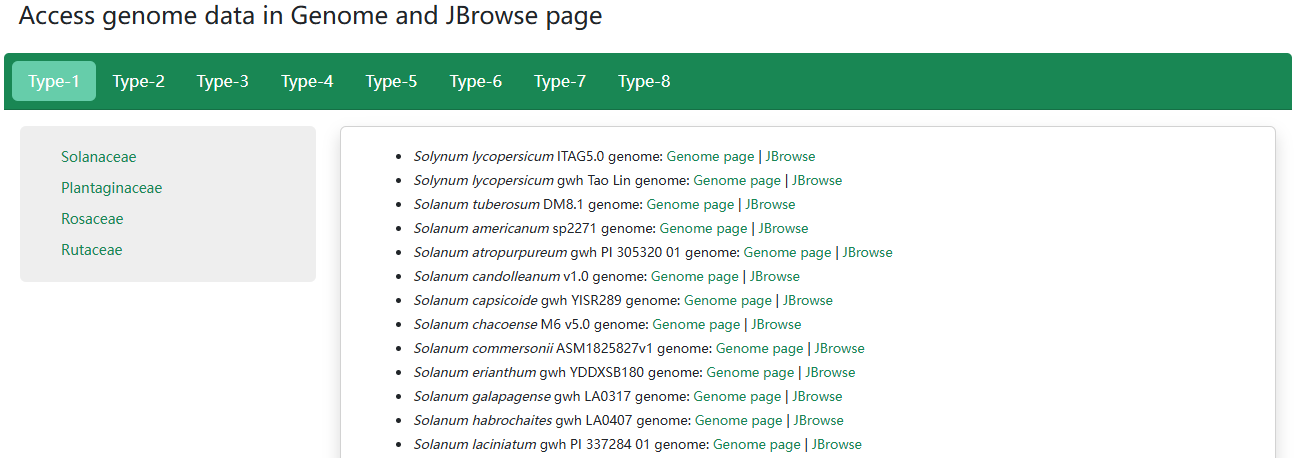
1. Access genome data in Genome and JBrowse page
The PSIA is the first comprehensive knowledgebase of plant SI, with all available genome assemblies of SI species collected from various public resources. In the genome browse page, users can take a quick browse on all genome assemblies in the current release, genome assemblies with the GFF file available will provide a Jbrowse function.
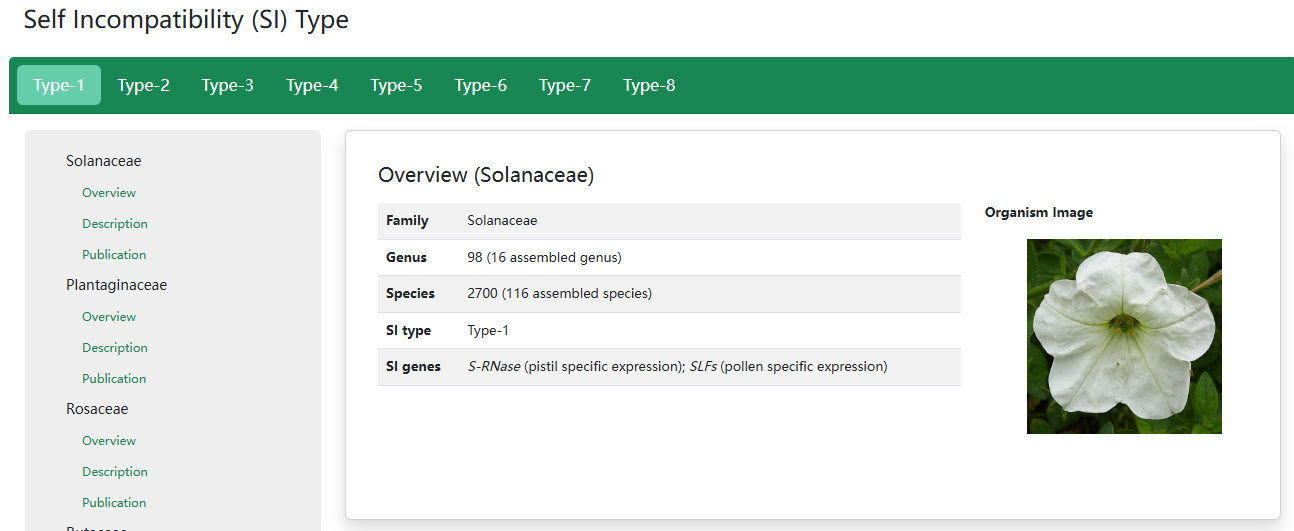
2. Self Incompatibility (SI) Type
In the previous studies of our group, we classified six molecular types of SI, including type-1 controlled by the pistil S S-RNase and pollen S SLFs commonly spread in Plantaginaceae, Solanaceae, Rosaceae and Rutaceae, type-2 by SRK and SCR in Brassicaceae, type-3 by PrsS and PrpS in Papaveraceae, type-4 by CYP-GLO2-KFB-CCM-PUM in Primulaceae, type-5 by SPH1-YUC6-BAHD in Turneraceae, type-6 by HPS10-S and DUF247I-S in Poaceae[ref1, 2]. Recently, we classified the SI of Linaceae as type-7 (two distyly candidate genes LtTSS1 and LtWDR-44 are only present in the dominant S allele) and classified the SI of Oleaceae as type-8 (the presence/absence polymorphism of the GA2ox-S gene is stably associated with SI groups across Oleaceae). The SI types were classified according to the molecular mechanism and S genes, and the serial number was based on the time the mechanism was discovered. In addition, we have summarized the molecular mechanisms of the eight SI types via cartoon diagrams.
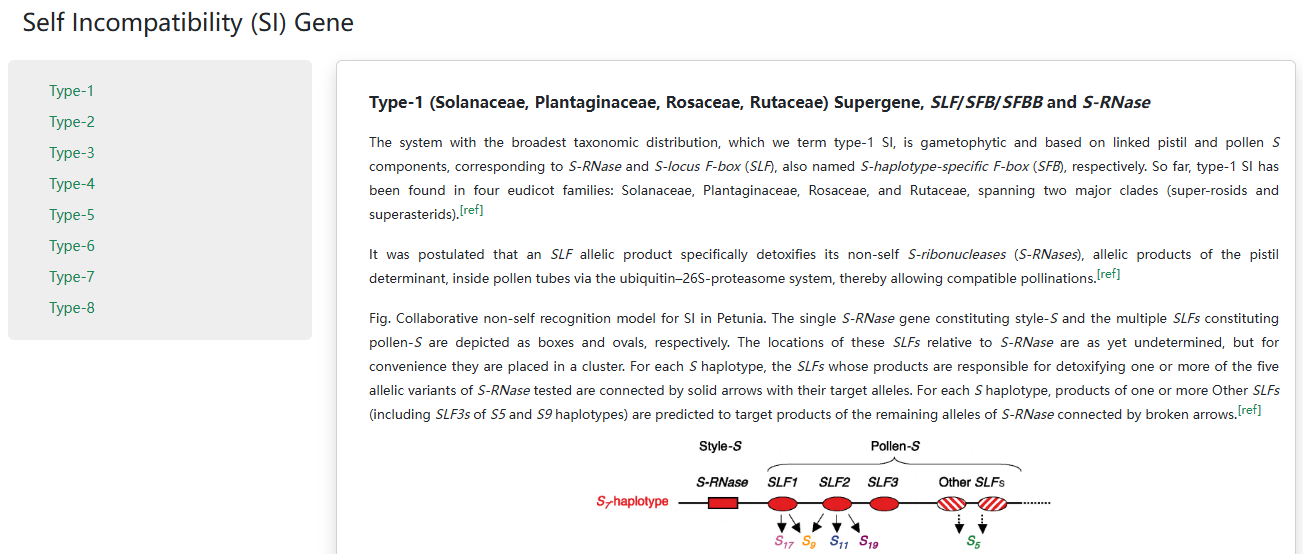
3. Self Incompatibility (SI) Gene
In the S-gene browse page, we introduced S genes of eight SI types. The genome organization or schematic diagram of S genes is provided by the pioneer research. The mechanism of SI genes is also shown in a part of SI types.
Species, Assemblies and S-locus Information
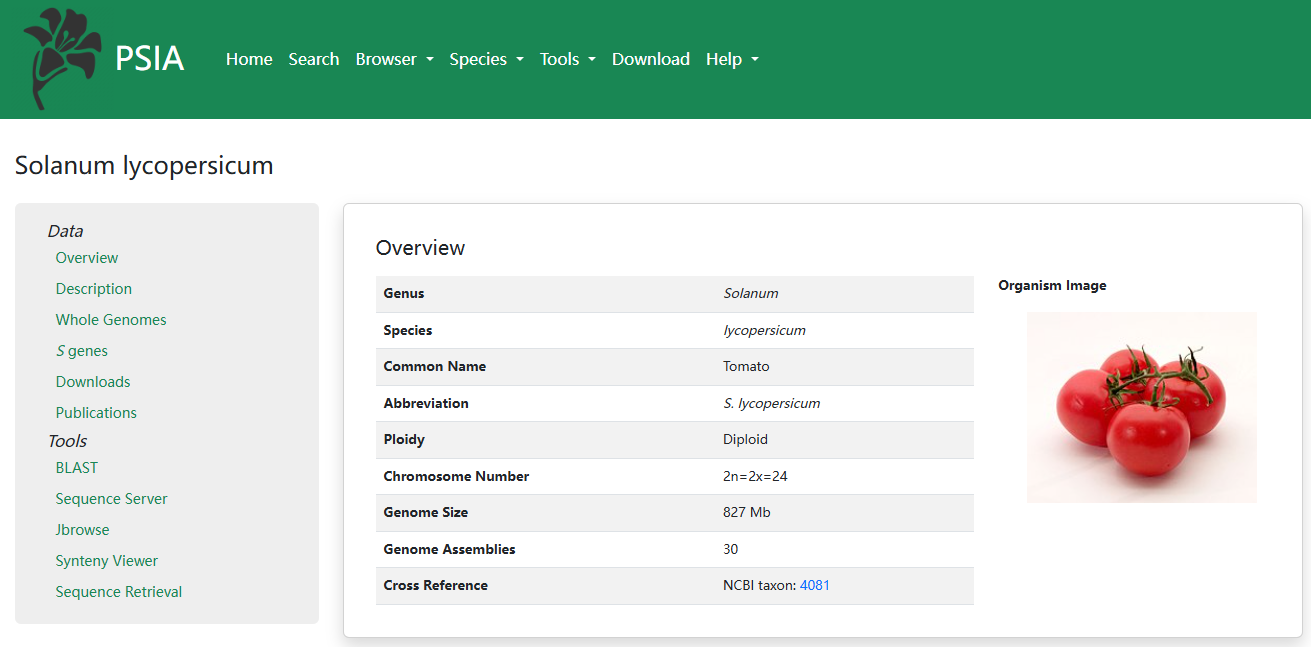
1. SI Species
PSIA provides an overview of each species. The total genome assembly counts are summarized from all available sources. On the left side, species description, genome assemblies page, S genes information, S genes download page and related publications are listed.
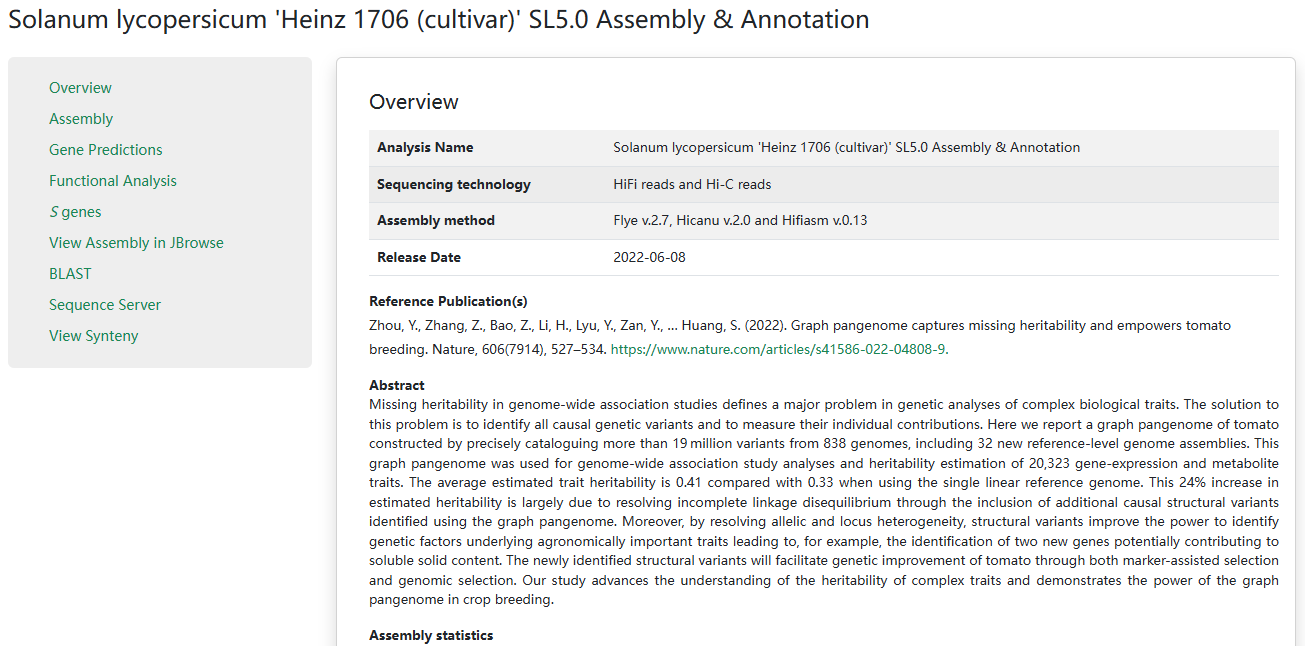
2. Genome assemblies
For each genome assembly, sequencing technology, assembly method and release date are collected. The reference, research abstract and assembly statistics are also provided. On the left side, we provide assembly, gene predictions, functional annotations and S-gene details.
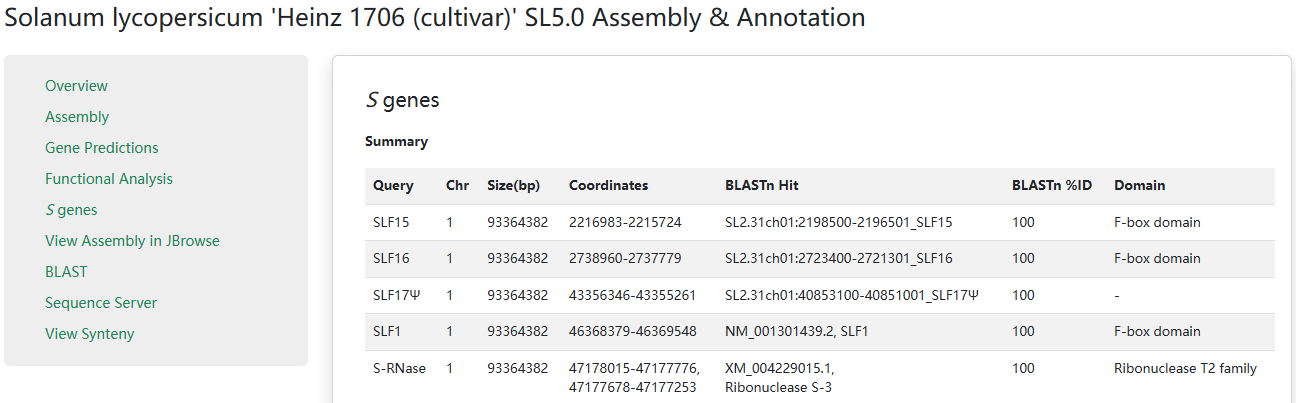
3. S-locus gene identification and curation
The S-locus information has a summary table and S genes list. The table gives users the genomic location and the BLAST hit of each gene. The conserved domains were identified using the NCBI Conserved Domain Database (CDD).
4. Curated S-locus gene in FASTA format
Each S-gene has been shown in FASTA format, and each fasta provides the basic information of the S gene, including genome assemblies, coordinates and blast hit.

Useful Tools
1. Workflow and results of BLAST in PSIA
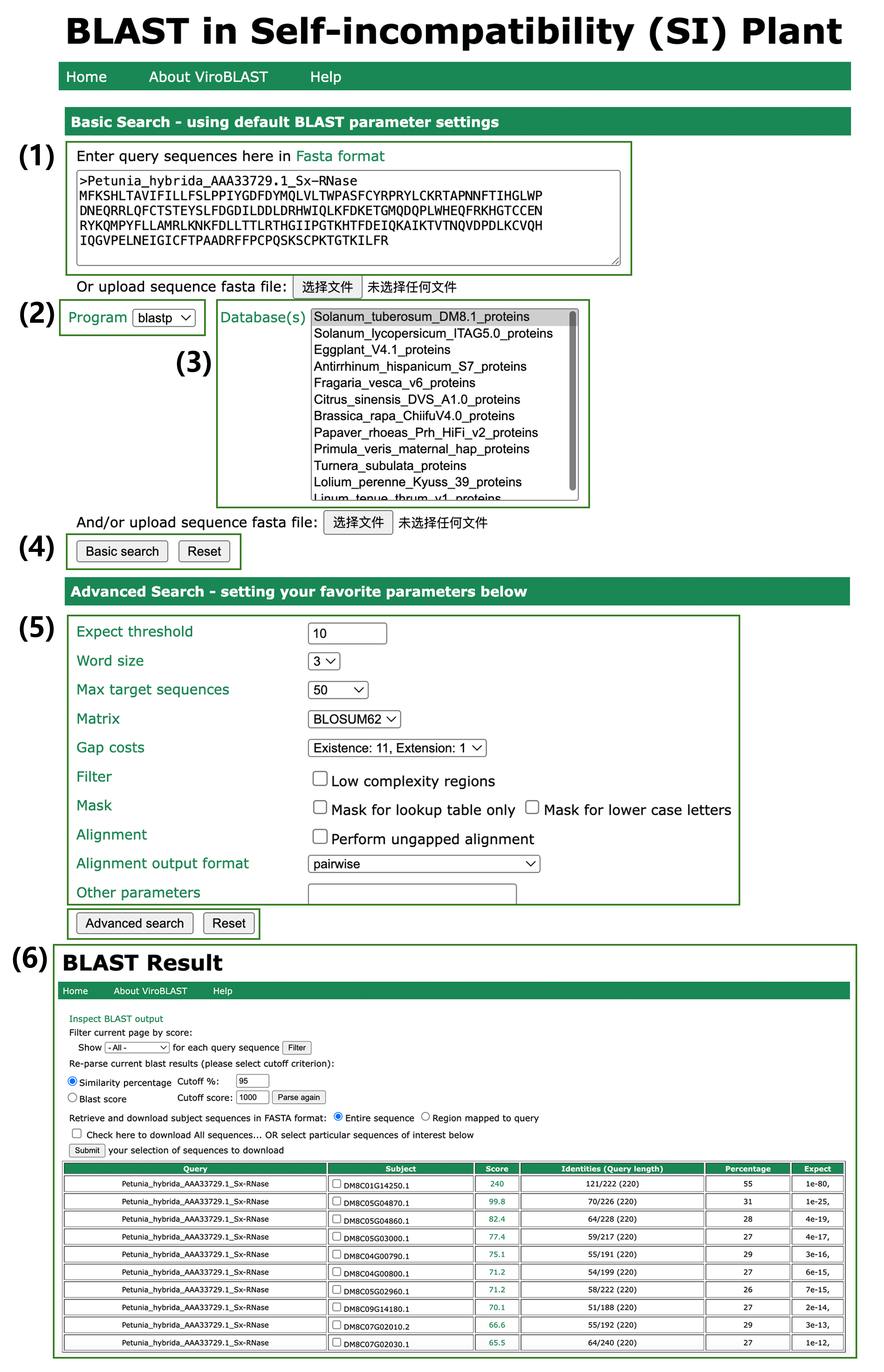
ViroBLAST has been integrated into PSIA, enhancing its BLAST functionality for genome-wide searches of self-incompatibility (SI) species. (1) Users can input a DNA or protein sequence. (2) Select the ‘blastn’ program for DNA sequences or ‘blastp’ for protein sequences. (3) Choose the appropriate BLAST local database for an SI species. (4) Obtain results using default parameters by clicking ‘Basic Search’. In most cases, the default parameters in ‘Basic Search’ yield optimal results. (5) For users requiring parameter customization, the ‘Advanced Search’ option allows fine-tuning of search settings before execution. (6) The BLAST results interface displays all matched alignments sorted by score. Results can be filtered based on similarity percentage or BLAST score.
2. Workflow and results of SequenceServer in PSIA.
SequenceServer has been integrated into PSIA, and local BLAST databases have been constructed using thousands of S gene sequences, including publicly known and newly identified sequences. (1) Users can input a DNA or protein sequence. (2) Select the appropriate BLAST local database of S genes. (3–4) Obtain results using default parameters. (5) The results page displays a graphical overview and a summary table of hits.


3. Example of JBrowse usage in PSIA
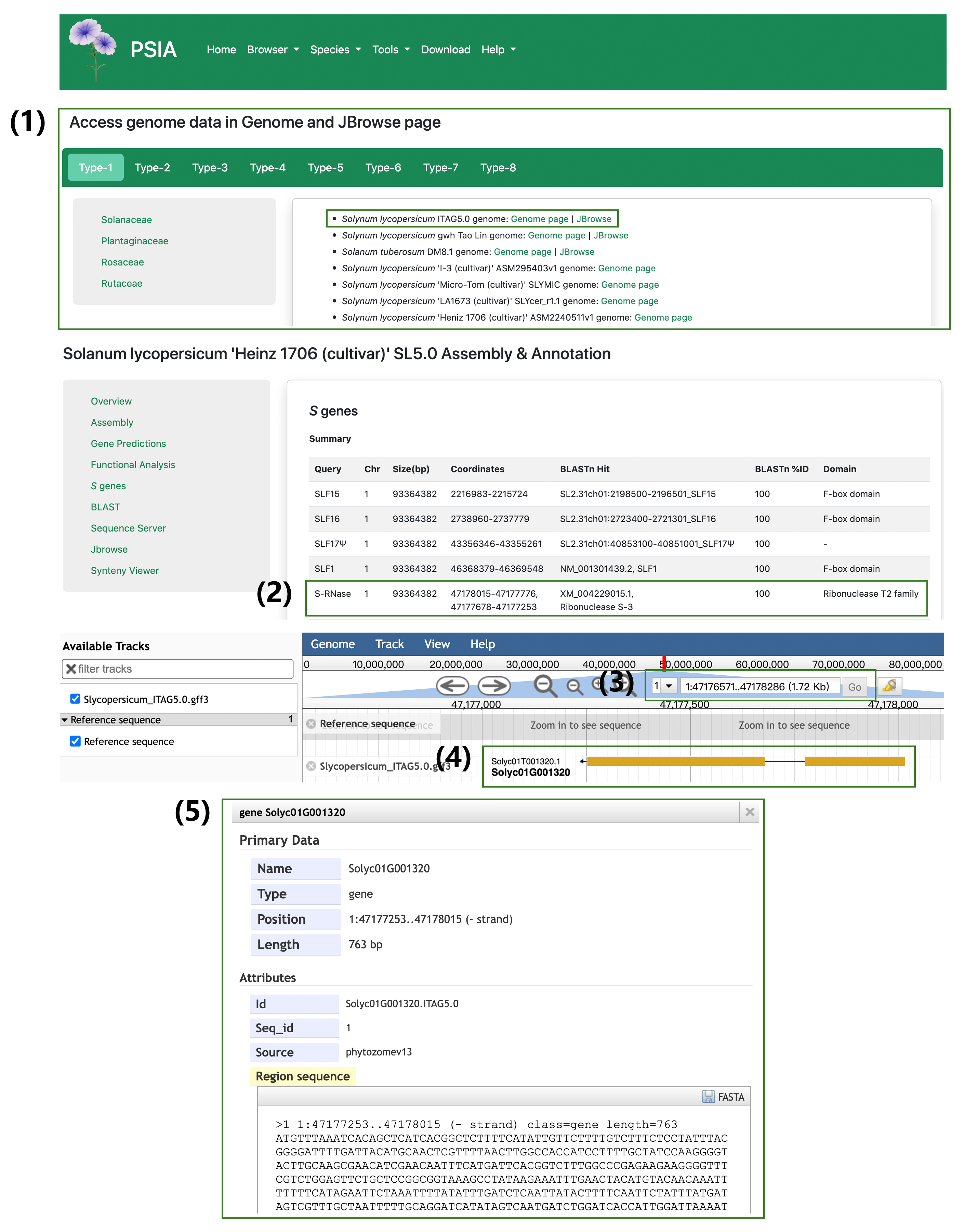
Genome assemblies with available GFF files provide JBrowse functionality, enabling researchers to explore the entire genome and S-locus. An example of JBrowse usage is as follows: (1) Select the JBrowse link for Solanum lycopersicum SL5.0. (2) View S-RNase gene information, including chromosomal location and genomic coordinates within the genome assembly. (3) In the JBrowse interface, enter the chromosome and genomic coordinates to visualize the gene structure of the S-RNase gene. (4–5) Click on the S-RNase gene to display detailed information about its structure and annotations.
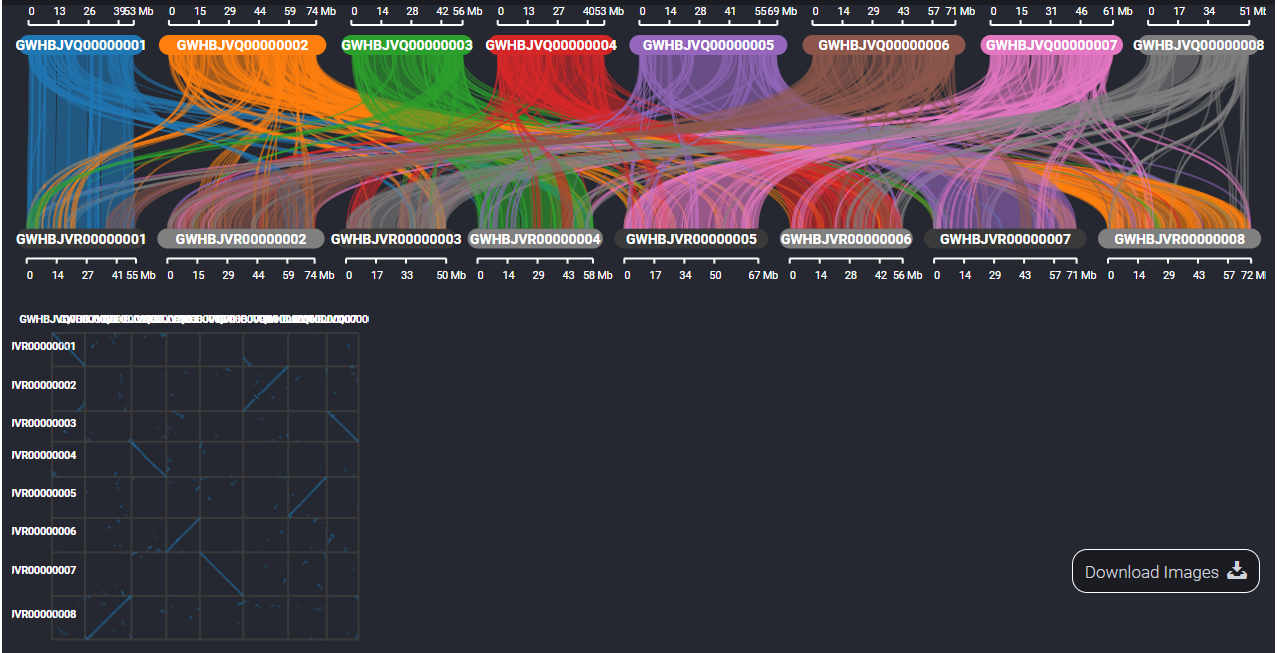
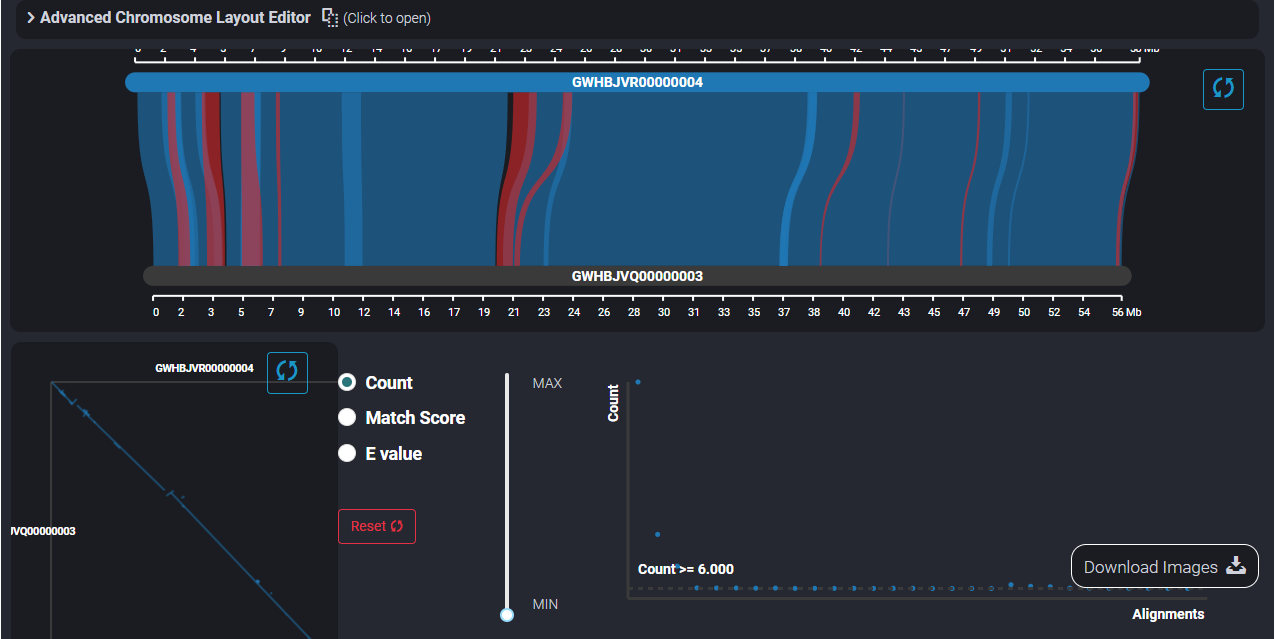
4. Workflow and example analysis results of Synteny Viewer
The SynVisio component will help users explore the result of comparative genomics between different genome assemblies. Here we show the result between Antirrhinum hispanicum S7 and A. hispanicum S8.
According to the S-locus location, we can compare the chromosomes of S-locus and find some pattern between them.
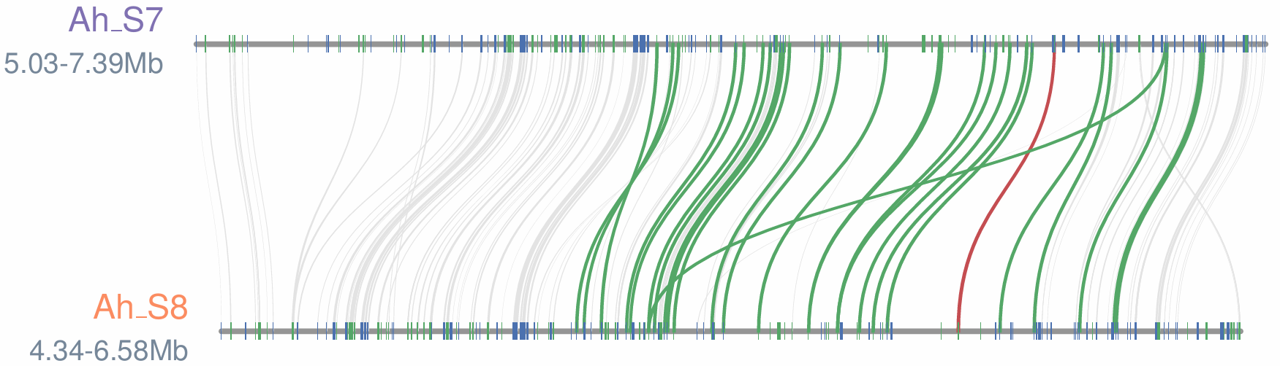
The S-locus synteny between A. hispanicum S7 and A. hispanicum S8 was generated using JCVI. The location of the S-RNase gene is indicated in red and the SLF genes are shown in green.
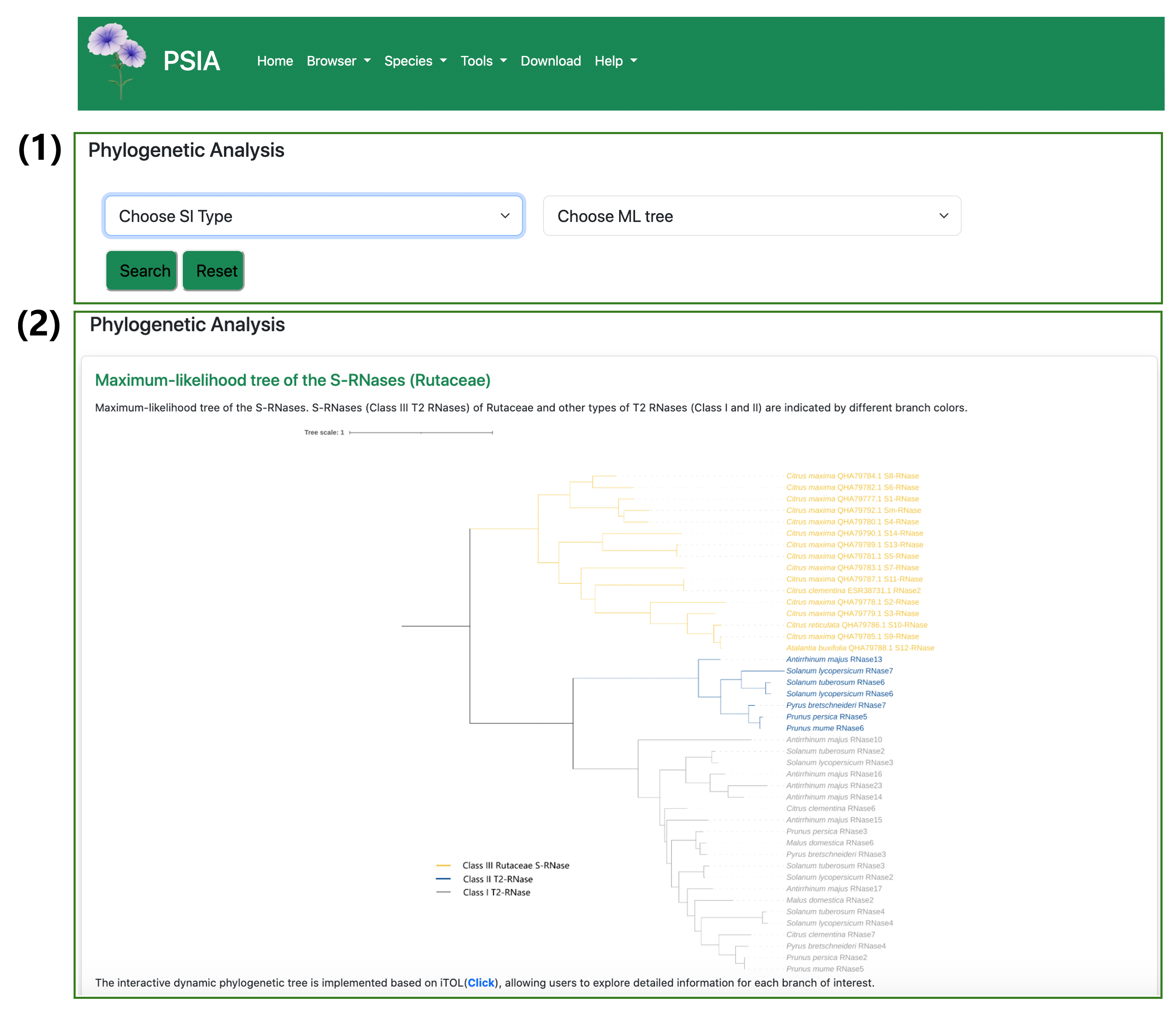
5. Workflow and example analysis results of Phylogenetic Analysis
(1) Using the Phylogenetic Analysis tool (accessed via the navigation bar), select the SI type-1 option in the search interface and choose ‘ML tree of S-RNases (Rutaceae)’ from the right-side dropdown menu. (2) The resulting phylogenetic tree displays evolutionary relationships among S-RNases (Class III T2 RNases) from Rutaceae and other T2 RNase classes (I and II).
6. User-friendly download
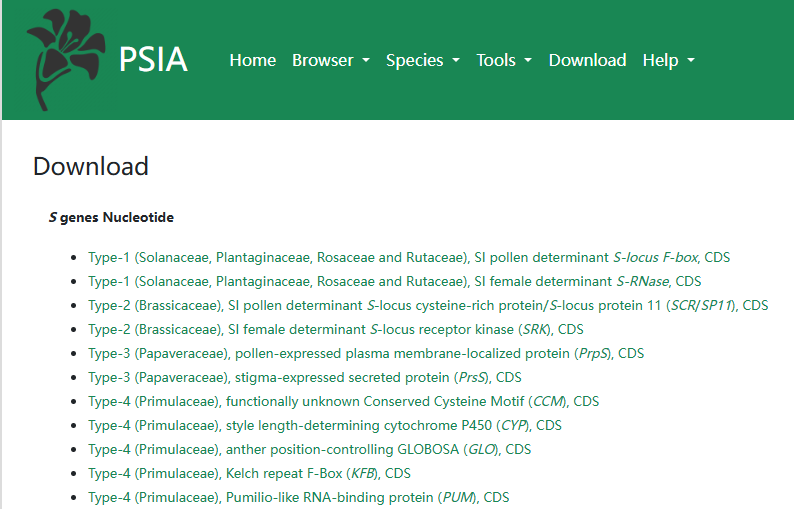
PSIA also provides a convenient download for all S genes, which can help biologist do their own specific research.