Rosaceae
Overview
| Family | Rosaceae |
| Genus | 91 (19 assembled genus) |
| Species | 4828 (74 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SFB/SFBB (pollen specific expression) |
Organism Image

Description
Rosaceae, the rose family, is a medium-sized family of flowering plants that includes 4,828 known species in 91 genera.
The name is derived from the type genus Rosa. The family includes herbs, shrubs, and trees. Most species are deciduous, but some are evergreen. They have a worldwide range but are most diverse in the Northern Hemisphere.
Many economically important products come from the Rosaceae, including various edible fruits, such as apples, pears, quinces, apricots, plums, cherries, peaches, raspberries, blackberries, loquats, strawberries, rose hips, hawthorns, and almonds. The family also includes popular ornamental trees and shrubs, such as roses, meadowsweets, rowans, firethorns, and photinias.
Among the most species-rich genera in the family are Alchemilla (270), Sorbus (260), Crataegus (260), Cotoneaster (260), Rubus (250), and Prunus (200), which contains the plums, cherries, peaches, apricots, and almonds. However, all of these numbers should be seen as estimates—much taxonomic work remains.
SI type
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.
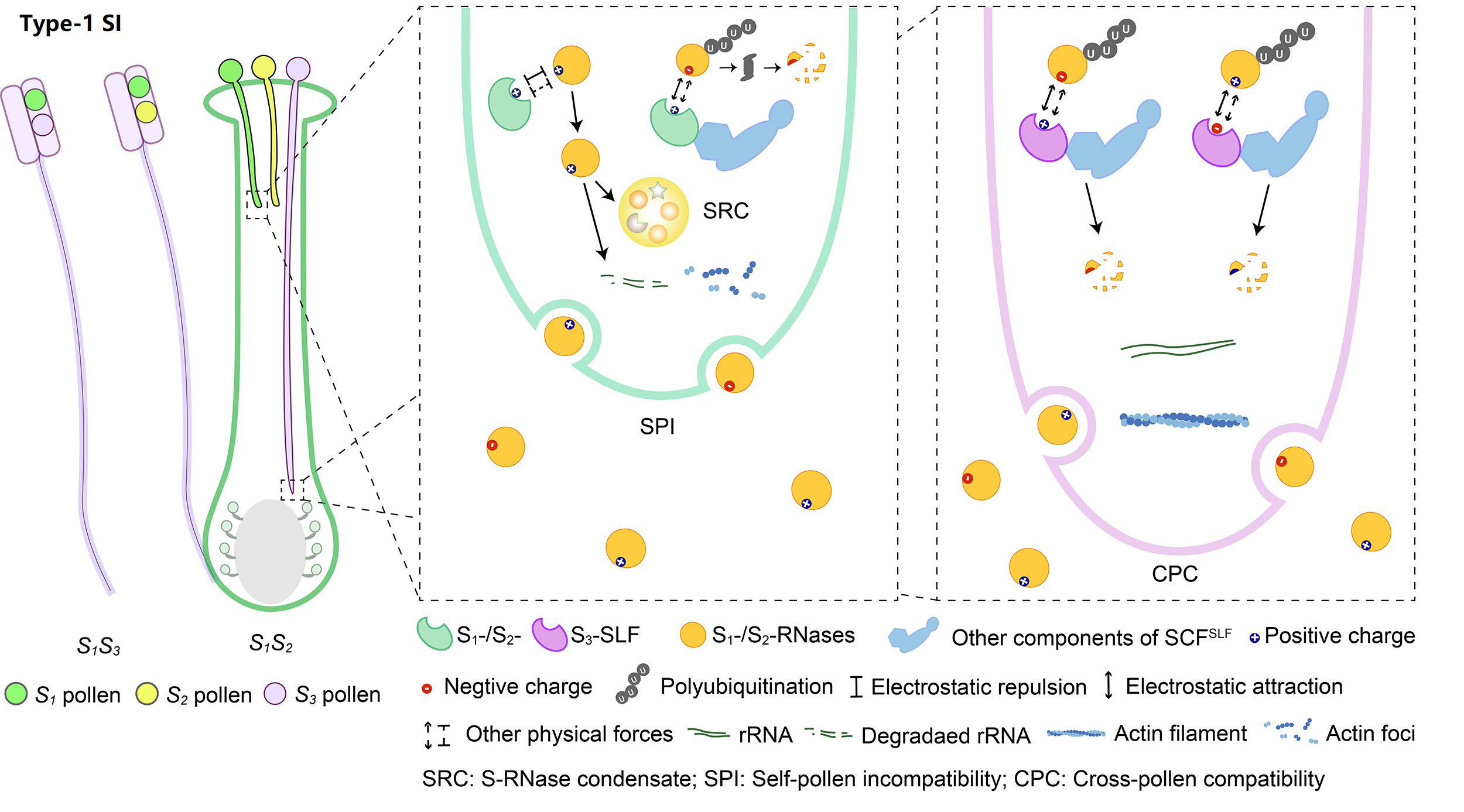
SI genes
Publication
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H. Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet. 1996 Mar 20;250(5):547-57. doi: 10.1007/BF02174443. Erratum in: Mol Gen Genet. 1996 Aug 27;252(1-2):222. doi: 10.1007/BF02173225.
Ishimizu T, Shinkawa T, Sakiyama F, Norioka S. Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Mol Biol. 1998 Aug;37(6):931-41. doi: 10.1023/a:1006078500664.
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell. 2003 Mar;15(3):771-81. doi: 10.1105/tpc.009290.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.