Prunus cerasus cv.Montmorency v1.0.a2 Assembly & Annotation
Overview
| Analysis Name | Prunus cerasus cv.Montmorency v1.0.a2 Assembly & Annotation |
| Sequencing technology | PacBio long-reads, Illumina short-reads |
| Assembly method | Canu |
| Release Date | 2024-02-21 |
Goeckeritz CZ, Rhoades KE, Childs KL, Iezzoni AF, VanBuren R, Hollender CA. Genome of tetraploid sour cherry (Prunus cerasus L.) 'Montmorency' identifies three distinct ancestral Prunus genomes. Hortic Res. 2023 May 10;10(7):uhad097. doi: 10.1093/hr/uhad097.
AbstractSour cherry (Prunus cerasus L.) is a valuable fruit crop in the Rosaceae family and a hybrid between progenitors closely related to extant Prunus fruticosa (ground cherry) and Prunus avium (sweet cherry). Here we report a chromosome-scale genome assembly for sour cherry cultivar Montmorency, the predominant cultivar grown in the USA. We also generated a draft assembly of P. fruticosa to use alongside a published P. avium sequence for syntelog-based subgenome assignments for 'Montmorency' and provide compelling evidence P. fruticosa is also an allotetraploid. Using hierarchal k-mer clustering and phylogenomics, we show 'Montmorency' is trigenomic, containing two distinct subgenomes inherited from a P. fruticosa-like ancestor (A and A') and two copies of the same subgenome inherited from a P. avium-like ancestor (BB). The genome composition of 'Montmorency' is AA'BB and little-to-no recombination has occurred between progenitor subgenomes (A/A' and B). In Prunus, two known classes of genes are important to breeding strategies: the self-incompatibility loci (S-alleles), which determine compatible crosses, successful fertilization, and fruit set, and the Dormancy Associated MADS-box genes (DAMs), which strongly affect dormancy transitions and flowering time. The S-alleles and DAMs in 'Montmorency' and P. fruticosa were manually annotated and support subgenome assignments. Lastly, the hybridization event 'Montmorency' is descended from was estimated to have occurred less than 1.61 million years ago, making sour cherry a relatively recent allotetraploid. The 'Montmorency' genome highlights the evolutionary complexity of the genus Prunus and will inform future breeding strategies for sour cherry, comparative genomics in the Rosaceae, and questions regarding neopolyploidy.
Assembly statistics
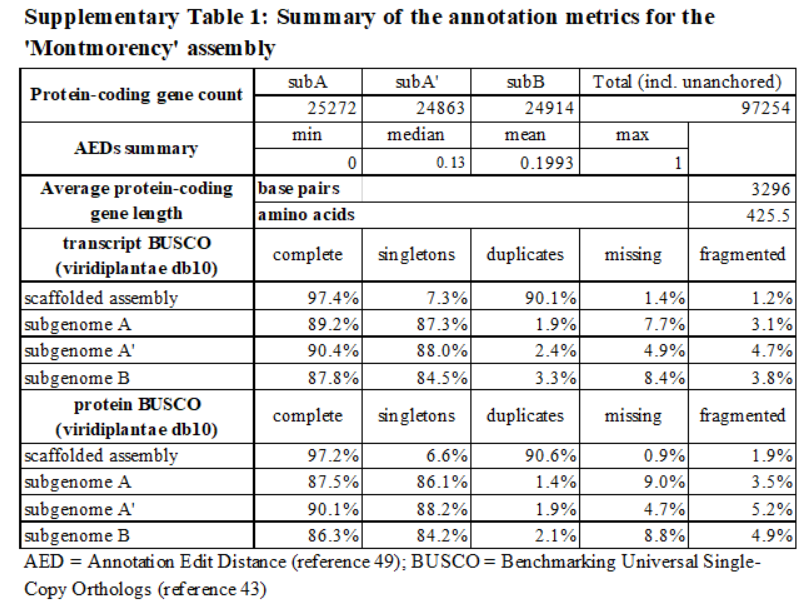
Assembly
The Prunus cerasus cv.Montmorency v1.0.a2 Assembly file is available in FASTA format.
Downloads
| Chromosomes (FASTA file) | Pcerasus_montmorency_final.fasta.gz |
Gene Predictions
The Prunus cerasus cv.Montmorency v1.0.a2 genome gene prediction files are available in GFF3 and FASTA format.
Downloads
| Genes (GFF3 file) | Pcer_mont_QTL_update2_CORRECTED.sortedb.gff3.gz |
| CDS sequences (FASTA file) | Pcer_mont_QTL_update2_cds.fasta.gz |
| Protein sequences (FASTA file) | Pcer_mont_QTL_update2_proteins.fasta.gz |
Functional Analysis
Functional annotation for the Prunus cerasus cv.Montmorency v1.0.a2 is available for download below. The proteins were analyzed using InterProScan to assign InterPro domains(Pfam).
Downloads
| Domain from InterProScan | Prunus_cerasus_cv.Montmorency_v1.0.a2.Pfam.tsv.gz |
S genes
Prunus S genes Nucleotide
- Prunus armeniaca S-locus F-box protein 4 (SFB4) gene, partial cds
- Prunus armeniaca S-locus SFBc (SFB) gene, complete cds
- Prunus armeniaca S14' F-box protein gene, complete cds
- Prunus armeniaca cultivar Xianhebao S-locus F-box protein 23 (SFB23) gene, complete cds
- Prunus armeniaca cultivar Jiguang S-locus F-box protein 8 gene, complete cds
- Prunus armeniaca S-locus F-box protein 33 (SFB33) mRNA, complete cds
- Prunus armeniaca S-locus F-box protein 50 (SFB50) mRNA, complete cds
- Prunus armeniaca S-haplotype-specific F-box protein (SFB) gene, SFB24 allele, complete cds
- Prunus armeniaca S-locus F-box protein 60 (SFB60) mRNA, complete cds
- Prunus armeniaca S-locus F-box protein-like (SFB) mRNA, complete sequence
- Prunus avium SFB3 mRNA for S-locus F-Box protein 3, complete cds
- Prunus avium SFB6 mRNA for S-locus F-Box protein 6, complete cds
- Prunus avium SFB1 gene for S haplotype-specific F-box protein 1, complete cds
- Prunus avium SFB2 gene for S haplotype-specific F-box protein 2, complete cds
- Prunus avium SFB5 gene for S haplotype-specific F-box protein 5, complete cds
- Prunus avium SFB4 mRNA for S haplotype-specific F-box protein 4, complete cds
- Prunus avium SLFL1-S4 gene for S locus F-box protein with the low allelic sequence polymorphism 1-S4, complete cds
- Prunus avium SLFL2-S4 gene for S locus F-box protein with the low allelic sequence polymorphism 2-S4, complete cds
- Prunus avium SLFL3-S4 gene for S locus F-box protein with the low allelic sequence polymorphism 3-S4, complete cds
- Prunus avium SLFL1-S1 gene for S locus F-box protein with the low allelic sequence polymorphism 1-S1, complete cds
- Prunus avium SLFL1-S2 gene for S locus F-box protein with the low allelic sequence polymorphism 1-S2, complete cds
- Prunus avium SLFL1-S5 gene for S locus F-box protein with the low allelic sequence polymorphism 1-S5, complete cds
- Prunus avium SFB3 (SFB) gene, SFB-S3 allele, complete cds
- Prunus avium S-locus F-box protein (SFB) gene, SFB-1 allele, partial cds
- Prunus avium S haplotype-specific F-box protein 13 (SFB) gene, SFB-S13 allele, complete cds
- Prunus avium S9-haplotype-specific F-box protein (SFB9) mRNA, SFB9-3 allele, complete cds
- Prunus avium S haplotype-specific F-box protein 7 (SFB) gene, SFB-S7 allele, complete cds
- Prunus avium cultivar Kronio truncated S-locus F-box protein (SFB) gene, SFB-S5' allele, complete cds
- Prunus avium F-box protein mRNA, complete cds
- Prunus avium SLFL6 mRNA for S locus F-box like protein 6, complete cds
- Prunus avium SLFL7A mRNA for S locus F-box like protein 7A, complete cds
- Prunus avium SFBL2 mRNA for S haplotype-specific F-box like protein 2, complete cds
- Prunus cerasus S haplotype-specific F-box protein 13' (SFB) gene, SFB-S13' allele, complete cds
- Prunus cerasus S haplotype-specific F-box protein 26 (SFB) gene, SFB-S26 allele, complete cds
- Prunus cerasus S haplotype-specific F-box protein 36a (SFB) gene, SFB-S36a allele, complete cds
- Prunus cerasus S haplotype-specific F-box protein 36b (SFB) gene, SFB-S36b allele, complete cds
- Prunus cerasus S haplotype-specific F-box protein 36b2 (SFB) gene, SFB-S36b2 allele, complete cds
- Prunus cerasus S haplotype-specific F-box protein 36b3 (SFB) gene, SFB-S36b3 allele, complete cds
- Prunus dulcis SFBc mRNA for S haplotype-specific F-box protein c, complete cds
- Prunus dulcis SFBd mRNA for S haplotype-specific F-box protein d, complete cds
- Prunus dulcis SFBa mRNA for S haplotype-specific F-box protein a, complete cds
- Prunus dulcis SFBb mRNA for S haplotype-specific F-box protein b, complete cds
- Prunus dulcis SLFc mRNA for S locus F-box protein c, complete cds
- Prunus dulcis SLFd mRNA for S locus F-box protein d, complete cds
- Prunus dulcis S locus F-box (SLF1) mRNA, complete cds
- Prunus dulcis S-haplotype-specific F-box protein (SFB) mRNA, SFB-S10 allele, complete cds
- Prunus dulcis cultivar Nonpareil S-haplotype-specific F-box protein 8 (SFB) gene, SFB-S8 allele, complete cds
- Prunus dulcis S-haplotype-specific F-box protein (SFB) mRNA, SFB-a allele, complete cds
- Prunus dulcis S-haplotype-specific F-box protein (SFB) mRNA, SFB-d allele, complete cds
- Prunus dulcis S-haplotype-specific F-box protein (SFB) mRNA, SFB-x allele, complete cds
- Prunus dulcis S-haplotype-specific F-box protein (SFB) mRNA, SFB-c allele, complete cds
- Prunus dulcis S-haplotype-specific F-box protein (SFB) mRNA, SFB-b allele, complete cds
- Prunus dulcis S-haplotype-specific F-box protein (SFB) mRNA, SFB-e allele, complete cds
- Prunus dulcis cultivar Dashitou S-haplotype-specific F-box protein (SFB) mRNA, SFB-g allele, complete cds
- Prunus dulcis cultivar Padre S haplotype-specific F-box protein (SFB) gene, SFB-18 allele, complete cds
- Prunus dulcis cultivar Ne Plus Ultra S haplotype-specific F-box protein (SFB) gene, SFB-7 allele, complete cds
- Prunus dulcis cultivar Mission S haplotype-specific F-box protein (SFB) gene, SFB-5 allele, complete cds
- Prunus dulcis cultivar Monterey S haplotype-specific F-box protein (SFB) gene, SFB-1 allele, complete cds
- Prunus dulcis cultivar Le Grand S haplotype-specific F-box protein (SFB) gene, SFB-1 allele, complete cds
- Prunus dulcis cultivar Ne Plus Ultra S haplotype-specific F-box protein (SFB) gene, SFB-1 allele, complete cds
- Prunus dulcis partial sfb gene for S-Locus F-Box protein, allele 11, cultivar selection ITAP-1, exon 1
- Prunus dulcis cultivar Nonpareil S locus F-box protein 8 (SLF8) gene, partial cds
- Prunus dulcis cultivar Nonpareil S haplotype-specific F-box protein 8 (SFB8) genes, complete cds
- Prunus dulcis cultivar McKinlays S haplotype-specific F-box protein 8 (SFB8) genes, complete cds
- Prunus dulcis cultivar McKinlays S locus F-box protein 8 (SLF8) complete cds
- Prunus dulcis cultivar Brown Nonpareil S haplotype-specific F-box protein 1 (SFB1) genes, complete cds
- Prunus dulcis cultivar Brown Nonpareil S locus F-box protein 1 (SLF1) gene, partial cds
- Prunus mume S1-SLF mRNA for F-box, complete cds
- Prunus mume S7-SLF mRNA for F-box, complete cds
- Prunus mume S7-SLFL1 gene for F-box, complete cds
- Prunus mume S7-SLFL2 gene for F-box, complete cds
- Prunus mume S7-SLFL3 gene for F-box, complete cds
- Prunus mume SFB1 mRNA for S-locus F-Box protein 1, complete cds
- Prunus mume SFB7 mRNA for S-locus F-Box protein 7, complete cds
- Prunus mume SLFL1-Sf gene for S locus F-box protein with the low allelic sequence polymorphism 1-Sf, complete cds
- Prunus mume SLFL2-Sf gene for S locus F-box protein with the low allelic sequence polymorphism 2-Sf, complete cds
- Prunus mume SLFL3-Sf gene for S locus F-box protein with the low allelic sequence polymorphism 3-Sf, complete cds
- Prunus mume F-box 1 gene, partial cds
- Prunus mume F-box 2 gene, partial cds
- PREDICTED: Prunus mume F-box/kelch-repeat protein At3g06240-like (LOC103343632), transcript variant X1, mRNA
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) mRNA, SFB-1 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) mRNA, SFB-5 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) mRNA, SFB-7 allele, complete cds
- Prunus pseudocerasus F-box protein (SFBc) mRNA, SFBc-s allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) gene, SFB11 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) gene, SFB12 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) gene, SFB13 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) gene, SFB14 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) mRNA, SFB-1 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) mRNA, SFB-4 allele, complete cds
- Prunus pseudocerasus S-haplotype-specific F-box protein (SFB) mRNA, SFB-6 allele, complete cds
- Prunus salicina F-box protein SFB (SFB) gene, SFB-c allele, complete cds
- Prunus salicina F-box protein SFB (SFB) gene, SFB-h allele, complete cds
- Prunus salicina cultivar black diamant S-locus-F-box protein e (SFB) gene, complete cds
- Prunus salicina cultivar stanley S-locus-F-box protein e (SFB) gene, complete cds
- Prunus salicina cultivar Santa Rosa S-locus-F-box protein b (SFB) gene, complete cds
- Prunus salicina cultivar bedri 1 S-locus-F-box protein c (SFB) gene, complete cds
- Prunus salicina cultivar jabounia safra S-locus-F-box protein h (SFB) gene, complete cds
- Prunus salicina cultivar janha S-locus-F-box protein f (SFB) gene, complete cds
- Prunus salicina cultivar cidre S-locus-F-box protein e (SFB) gene, complete cds
- Prunus salicina cultivar aouina hamra bedria S-locus-F-box protein e (SFB) gene, complete cds
- Prunus salicina cultivar ain torkia S-locus-F-box protein c (SFB) gene, complete cds
- Prunus salicina cultivar meski hamra S-locus-F-box protein f (SFB) gene, complete cds
- Prunus salicina cultivar zaghwenia S-locus-F-box protein f (SFB) gene, complete cds
- Prunus salicina cultivar ain tasstouria S-locus-F-box protein h (SFB) gene, complete cds
- Prunus salicina cultivar meski kahla S-locus-F-box protein f (SFB) gene, complete cds
- Prunus salicina cultivar hamra bedri S-locus-F-box protein b (SFB) gene, complete cds
- Prunus salicina cultivar bedri2 S-locus-F-box protein h (SFB) gene, complete cds
- Prunus salicina cultivar black golden S-locus-F-box protein b (SFB) gene, complete cds
- Prunus speciosa S-haplotype-specific F-box protein (SFB) gene, SFB1 allele, complete cds
- Prunus speciosa S-haplotype-specific F-box protein (SFB) gene, SFB22 allele, complete cds
- Prunus speciosa S-haplotype-specific F-box protein (SFB) gene, SFB31 allele, complete cds
- Prunus speciosa S-haplotype-specific F-box protein (SFB) gene, SFB51 allele, complete cds
- Prunus tenella S-locus F-box protein (S8_SFB) gene, partial cds
- Prunus tenella SFB16 protein mRNA, complete cds
- Prunus tenella SFB17 protein mRNA, complete cds
- Prunus webbii S locus F-box protein (SLF) gene, partial cds
- Prunus armeniaca S1-RNase protein (S1-RNase) gene, partial cds
- Prunus armeniaca S4-RNase protein (S4-RNase) gene, partial cds
- Prunus armeniaca S-locus S-RNase c (S-RNase) gene, complete cds
- Prunus armeniaca S-locus associated ribonuclease (S-RNase) gene, S-RNase-S24 allele, complete cds
- Prunus armeniaca S-locus associated ribonuclease (S-RNase) gene, S-RNase-S24 allele, complete cds
- Prunus armeniaca S locus S-RNase 52 (S-RNase) gene, complete cds
- Prunus armeniaca S-RNase 53 (S-RNase) gene, complete cds
- Prunus armeniaca mRNA for S30-RNase, complete cds
- Prunus armeniaca mRNA for S31-RNase, complete cds
- Prunus avium mRNA for S4-RNase, complete cds
- Prunus avium mRNA for RNase S1 (S1 gene)
- Prunus avium S-RNase (S) gene, S24 allele, complete cds
- Prunus avium S-RNase (S) gene, S25 allele, complete cds
- Prunus avium S-RNase (S) gene, S23 allele, complete cds
- Prunus avium S-RNase (S) gene, S12 allele, complete cds
- Prunus avium S13-RNase (S-RNase) gene, S-RNase-S13 allele, complete cds
- Prunus avium S7-RNase (S-RNase) gene, S-RNase-S7 allele, complete cds
- Prunus avium cultivar Techlovicka S-RNase S54 (S) gene, S54 allele, complete cds
- Prunus cerasus S6m2-RNase (S-RNase) gene, S-RNase-S6m2 allele, complete cds
- Prunus cerasus S13m-RNase (S-RNase) gene, S-RNase-S13m allele, complete cds
- Prunus cerasus S26-RNase (S-RNase) gene, S-RNase-S26 allele, complete cds
- Prunus cerasus S36a-RNase (S-RNase) gene, S-RNase-S36a allele, complete cds
- Prunus cerasus S36b-RNase (S-RNase) gene, S-RNase-S36b allele, complete cds
- Prunus cerasus S36b3-RNase (S-RNase) gene, S-RNase-S36b3 allele, complete cds
- Prunus cerasus cultivar Meteor S33-RNase gene, complete cds
- Prunus cerasus S34-RNase gene, complete cds
- Prunus dulcis mRNA for Sb-RNase, complete cds
- Prunus dulcis mRNA for Sc-RNase, complete cds
- Prunus dulcis mRNA for Sa-RNase, complete cds
- Prunus dulcis mRNA for Sa-RNase, complete cds
- Prunus dulcis S-RNase gene for S30-RNase, complete cds
- Prunus dulcis S-RNase gene for Sf-RNase, complete cds
- Prunus dulcis S gene for S23-RNase, complete cds
- Prunus dulcis S-RNase (Sn) mRNA, complete cds
- Prunus dulcis S-RNase (Sm) mRNA, complete cds
- Prunus dulcis cultivar Tuono S-RNase (S) mRNA, Sf allele, complete cds
- Prunus dulcis cultivar Nonpareil S8-RNase, complete cds
- Prunus dulcis cultivar McKinlays S8-RNase, complete cds
- Prunus dulcis cultivar Brown Nonpareil S1-RNase, complete cds
- Prunus mume S7-RNase mRNA for S7-ribonuclease, partial cds
- Prunus mume mRNA for Sf-RNase, complete cds
- Prunus mume mRNA for S1-RNase, complete cds
- Prunus mume S-RNase gene for S-ribonuclease, complete cds, allele: S1
- Prunus mume S-RNase gene for S-ribonuclease, complete cds, allele: S7
- Prunus mume S14-RNase mRNA, complete cds
- Prunus mume S15-RNase mRNA, complete cds
- Prunus mume S16-RNase mRNA, complete cds
- PREDICTED: Prunus mume ribonuclease MC-like (LOC103343622), mRNA
- PREDICTED: Prunus persica ribonuclease MC (LOC18772864), mRNA
- PREDICTED: Prunus persica ribonuclease S-2 (LOC18769002), mRNA
- Prunus pseudocerasus S-RNase (S1-RNase) mRNA, S1-RNase-S allele, complete cds
- Prunus pseudocerasus S-RNase (S2-RNase) mRNA, S2-RNase-S allele, complete cds
- Prunus pseudocerasus self-incompatibility associated ribonuclease S2 (S-RNase) mRNA, S-RNase-S2 allele, complete cds
- Prunus pseudocerasus self-incompatibility associated ribonuclease S4 (S-RNase) mRNA, S-RNase-S4 allele, complete cds
- Prunus pseudocerasus self-incompatibility associated ribonuclease S6 (S-RNase) mRNA, S-RNase-S6 allele, complete cds
- Prunus pseudocerasus S-locus-associated ribonuclease (S-RNase) gene, S-RNase-S11 allele, complete cds
- Prunus pseudocerasus S-locus-associated ribonuclease (S-RNase) gene, S-RNase-S12 allele, complete cds
- Prunus pseudocerasus S-locus-associated ribonuclease (S-RNase) gene, S-RNase-S13 allele, complete cds
- Prunus pseudocerasus self-incompatibility associated ribonuclease S1 (S-RNase) gene, S-RNase-S1 allele, complete cds
- Prunus pseudocerasus self-incompatibility associated ribonuclease S5 (S-RNase) gene, S-RNase-S5 allele, complete cds
- Prunus pseudocerasus self-incompatibility associated ribonuclease S8 (S-RNase) mRNA, S-RNase-S8 allele, complete cds
- Prunus salicina PS-Sc mRNA for Sc-RNase, complete cds
- Prunus salicina PS-Sc mRNA for Sc-RNase, complete cds
- Prunus salicina PS-Sd mRNA for Sd-RNase, complete cds
- Prunus salicina gene for Sb-RNase, complete cds
- Prunus speciosa S-locus-associated ribonuclease (S-RNase) gene, S-RNase-S1 allele, complete cds
- Prunus speciosa S-locus-associated ribonuclease (S-RNase) gene, S-RNase-S31 allele, complete cds
- Prunus speciosa S-locus-associated ribonuclease (S-RNase) gene, S-RNase-S51 allele, complete cds
- Prunus speciosa S-locus associated ribonuclease (S-RNase) gene, S-RNase-S22 allele, complete cds
- Prunus spinosa S-locus S-RNase S16 gene, complete cds
- Prunus subhirtella S gene for S-RNase, complete cds, allele: S1
- Prunus tenella S12 S-RNase mRNA, complete cds
- Prunus tenella S14 S-RNase mRNA, complete cds
- Prunus tenella S16-RNase protein mRNA, complete cds
- Prunus tenella S17-RNase protein mRNA, complete cds
- Prunus webbii S-RNase (S2) mRNA, complete cds
- Prunus x yedoensis S mRNA for S-RNase, complete cds, allele: PyS1
- Prunus x yedoensis S mRNA for S-RNase, complete cds, allele: PyS2
- Prunus x yedoensis S gene for S-RNase, complete cds, allele: PyS1
- Prunus x yedoensis S gene for S-RNase, complete cds, allele: PyS2
Prunus S genes Protein
- S-locus F-box protein 4 [Prunus armeniaca]
- S-locus SFBc [Prunus armeniaca]
- S14' F-box protein [Prunus armeniaca]
- S-locus F-box protein 23 [Prunus armeniaca]
- S-locus F-box protein 8 [Prunus armeniaca]
- S-locus F-box protein 33 [Prunus armeniaca]
- S-locus F-box protein 50 [Prunus armeniaca]
- S-haplotype-specific F-box protein [Prunus armeniaca], allele SFB24
- S-locus F-box protein 60 [Prunus armeniaca]
- SFB3 [Prunus avium]
- S-locus F-box protein, partial [Prunus avium], allele 1
- S haplotype-specific F-box protein 13 [Prunus avium]
- S9-haplotype-specific F-box protein [Prunus avium]
- S haplotype-specific F-box protein 7 [Prunus avium]
- truncated S-locus F-box protein [Prunus avium], allele S5'
- F-box protein [Prunus avium], PavFB
- S-locus F-Box protein 3 [Prunus avium]
- S-locus F-Box protein 6 [Prunus avium]
- S haplotype-specific F-box protein 1 [Prunus avium]
- S haplotype-specific F-box protein 2 [Prunus avium]
- S haplotype-specific F-box protein 5 [Prunus avium]
- S haplotype-specific F-box protein 4 [Prunus avium]
- S locus F-box protein with the low allelic sequence polymorphism 1-S4 [Prunus avium]
- S locus F-box protein with the low allelic sequence polymorphism 2-S4 [Prunus avium]
- S locus F-box protein with the low allelic sequence polymorphism 3-S4 [Prunus avium]
- S locus F-box protein with the low allelic sequence polymorphism 1-S1 [Prunus avium]
- S locus F-box protein with the low allelic sequence polymorphism 1-S2 [Prunus avium]
- S locus F-box protein with the low allelic sequence polymorphism 1-S5 [Prunus avium]
- S locus F-box like protein 6 [Prunus avium]
- S locus F-box like protein 7A [Prunus avium]
- S haplotype-specific F-box like protein 2 [Prunus avium]
- S haplotype-specific F-box protein 13' [Prunus cerasus]
- S haplotype-specific F-box protein 26 [Prunus cerasus]
- S haplotype-specific F-box protein 36a [Prunus cerasus]
- S haplotype-specific F-box protein 36b [Prunus cerasus]
- S haplotype-specific F-box protein 36b2 [Prunus cerasus]
- S haplotype-specific F-box protein 36b3 [Prunus cerasus]
- S locus F-box [Prunus dulcis], gene SLF1
- S-haplotype-specific F-box protein [Prunus dulcis], allele SFB-S10
- S-haplotype-specific F-box protein 8 [Prunus dulcis]
- S-haplotype-specific F-box protein [Prunus dulcis], allele a
- S-haplotype-specific F-box protein [Prunus dulcis], allele d
- S-haplotype-specific F-box protein [Prunus dulcis], allele x
- S-haplotype-specific F-box protein [Prunus dulcis], allele c
- S-haplotype-specific F-box protein [Prunus dulcis], allele b
- S-haplotype-specific F-box protein [Prunus dulcis], allele e
- S haplotype-specific F-box protein [Prunus dulcis], allele 18
- S haplotype-specific F-box protein [Prunus dulcis], allele 7
- S haplotype-specific F-box protein [Prunus dulcis], allele 5
- S haplotype-specific F-box protein [Prunus dulcis], allele 1
- S haplotype-specific F-box protein [Prunus dulcis], allele 1
- S haplotype-specific F-box protein [Prunus dulcis], allele 1
- S haplotype-specific F-box protein c [Prunus dulcis]
- S haplotype-specific F-box protein d [Prunus dulcis]
- S haplotype-specific F-box protein a [Prunus dulcis]
- S haplotype-specific F-box protein b [Prunus dulcis]
- S haplotype-specific F-box protein c [Prunus dulcis]
- S haplotype-specific F-box protein d [Prunus dulcis]
- S-Locus F-Box protein, partial [Prunus dulcis], allele 11
- S-haplotype-specific F-box protein [Prunus dulcis], allele g
- S locus F-box protein 8, partial [Prunus dulcis]
- S haplotype-specific F-box protein 8 [Prunus dulcis]
- S locus F-box protein 8 [Prunus dulcis]
- S haplotype-specific F-box protein 8 [Prunus dulcis]
- S locus F-box protein 1, partial [Prunus dulcis]
- S haplotype-specific F-box protein 1 [Prunus dulcis]
- F-box 1, partial [Prunus mume]
- F-box 2, partial [Prunus mume]
- F-box [Prunus mume], S1-SLF
- F-box [Prunus mume], S7-SLF
- F-box [Prunus mume], S7-SLFL1
- F-box [Prunus mume], S7-SLFL2
- F-box [Prunus mume], S7-SLFL3
- S-locus F-Box protein 1 [Prunus mume]
- S-locus F-Box protein 7 [Prunus mume]
- S locus F-box protein with the low allelic sequence polymorphism 1-Sf [Prunus mume]
- S locus F-box protein with the low allelic sequence polymorphism 2-Sf [Prunus mume]
- S locus F-box protein with the low allelic sequence polymorphism 3-Sf [Prunus mume]
- PREDICTED: F-box/kelch-repeat protein At3g06240-like isoform X1 [Prunus mume]
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB-1
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB-5
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB-7
- F-box protein [Prunus pseudocerasus], SFBc
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB-1
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB-4
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB-6
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB11
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB12
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB13
- S-haplotype-specific F-box protein [Prunus pseudocerasus], SFB14
- F-box protein SFB [Prunus salicina], allele c
- F-box protein SFB [Prunus salicina], allele h
- S-locus-F-box protein e [Prunus salicina], SFB
- S-locus-F-box protein e [Prunus salicina]
- S-locus-F-box protein b [Prunus salicina]
- S-locus-F-box protein c [Prunus salicina]
- S-locus-F-box protein h [Prunus salicina]
- S-locus-F-box protein f [Prunus salicina]
- S-locus-F-box protein e [Prunus salicina]
- S-locus-F-box protein e [Prunus salicina]
- S-locus-F-box protein c [Prunus salicina]
- S-locus-F-box protein f [Prunus salicina]
- S-locus-F-box protein f [Prunus salicina]
- S-locus-F-box protein h [Prunus salicina]
- S-locus-F-box protein f [Prunus salicina]
- S-locus-F-box protein b [Prunus salicina]
- S-locus-F-box protein h [Prunus salicina]
- S-locus-F-box protein b [Prunus salicina]
- S-haplotype-specific F-box protein [Prunus speciosa], SFB1
- S-haplotype-specific F-box protein [Prunus speciosa], SFB22
- S-haplotype-specific F-box protein [Prunus speciosa], SFB31
- S-haplotype-specific F-box protein [Prunus speciosa], SFB51
- S-locus F-box protein, partial [Prunus tenella], S8_SFB
- SFB16 protein [Prunus tenella]
- SFB17 protein [Prunus tenella]
- S locus F-box protein, partial [Prunus webbii]
- S1-RNase protein [Prunus armeniaca]
- S4-RNase protein [Prunus armeniaca]
- S-locus S-RNase c [Prunus armeniaca]
- S-locus associated ribonuclease [Prunus armeniaca], allele S24
- S-locus associated ribonuclease [Prunus armeniaca], allele S24
- S locus S-RNase 52 [Prunus armeniaca]
- S-RNase 53 [Prunus armeniaca]
- S30-RNase [Prunus armeniaca]
- S31-RNase [Prunus armeniaca]
- S-RNase [Prunus avium], allele S24
- S-RNase [Prunus avium], allele S25
- S-RNase [Prunus avium], allele S23
- S-RNase [Prunus avium], allele S12
- S13-RNase [Prunus avium]
- S7-RNase [Prunus avium]
- S4-RNase [Prunus avium]
- RNase S1 [Prunus avium]
- S-RNase S54 [Prunus avium]
- S6m2-RNase [Prunus cerasus]
- S13m-RNase [Prunus cerasus]
- S26-RNase [Prunus cerasus]
- S36a-RNase [Prunus cerasus]
- S36b-RNase [Prunus cerasus]
- S36b3-RNase [Prunus cerasus]
- S33-RNase [Prunus cerasus]
- S34-RNase [Prunus cerasus]
- S-RNase [Prunus dulcis], gene Sn
- S-RNase [Prunus dulcis], gene Sm
- S-RNase [Prunus dulcis], allele Sf
- Sb-RNase [Prunus dulcis]
- Sc-RNase [Prunus dulcis]
- Sa-RNase [Prunus dulcis]
- S30-RNase [Prunus dulcis]
- Sf-RNase [Prunus dulcis]
- S23-RNase [Prunus dulcis]
- S8-RNase [Prunus dulcis]
- S8-RNase [Prunus dulcis]
- S1-RNase [Prunus dulcis]
- S14-RNase [Prunus mume]
- S15-RNase [Prunus mume]
- S16-RNase [Prunus mume]
- Sf-RNase [Prunus mume]
- S1-RNase [Prunus mume]
- S7-ribonuclease, partial [Prunus mume]
- S-ribonuclease [Prunus mume], allele S1
- S-ribonuclease [Prunus mume], allele S7
- PREDICTED: ribonuclease MC-like [Prunus mume]
- ribonuclease MC [Prunus persica]
- ribonuclease S-2 [Prunus persica]
- S-RNase [Prunus pseudocerasus], S1-RNase
- S-RNase [Prunus pseudocerasus], S2-RNase
- self-incompatibility associated ribonuclease S2 [Prunus pseudocerasus]
- self-incompatibility associated ribonuclease S4 [Prunus pseudocerasus]
- self-incompatibility associated ribonuclease S6 [Prunus pseudocerasus]
- S-locus-associated ribonuclease [Prunus pseudocerasus], allele S11
- S-locus-associated ribonuclease [Prunus pseudocerasus], allele S12
- S-locus-associated ribonuclease [Prunus pseudocerasus], allele S13
- self-incompatibility associated ribonuclease S1 [Prunus pseudocerasus], allele S1
- self-incompatibility associated ribonuclease S5 [Prunus pseudocerasus]
- self-incompatibility associated ribonuclease S8 [Prunus pseudocerasus]
- Sc-RNase [Prunus salicina]
- Sb-RNase [Prunus salicina]
- S-locus-associated ribonuclease [Prunus speciosa], allele S1
- S-locus-associated ribonuclease [Prunus speciosa], allele S31
- S-locus-associated ribonuclease [Prunus speciosa], allele S51
- S-locus associated ribonuclease [Prunus speciosa], allele S22
- S-locus S-RNase S16 [Prunus spinosa]
- S-RNase [Prunus subhirtella var. ascendens], allele S1
- S12 S-RNase [Prunus tenella]
- S14 S-RNase [Prunus tenella]
- S16-RNase protein [Prunus tenella]
- S17-RNase protein [Prunus tenella]
- S-RNase [Prunus webbii]
- S-RNase [Prunus yedoensis], allele PyS1
- S-RNase [Prunus yedoensis], allele PyS2
- S-RNase [Prunus yedoensis], allele PyS1
- S-RNase [Prunus yedoensis], allele PyS2