Origin and evolution of plant SI
In the five types of self-incompatibility (SI) systems identified in eudicots, type 1 SI is the oldest and most widespread, while types 2 to 5 are specific to families such as Brassicaceae, Papaveraceae, Primulaceae, and Passifloraceae. Xue et al. were pioneers in suggesting that type 1 SI has a single origin[ref1]. The T2 RNases related to the S factor of type 1 styles fall into three branches, labeled class I, II, and III, with S-RNase forming a distinct branch within class III. Beyond the families known to have type 1 SI systems (such as Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae), T2 nucleases from other families such as Malvaceae, Rubiaceae, Euphorbiaceae, Cucurbitaceae, Fabaceae, Oleaceae, Theaceae, and Rhamnaceae can also be grouped with class III T2 nucleases/S-RNase. This grouping includes only eudicots, suggesting that the S-RNase of type 1 has a single origin from the most recent common ancestor(MRCA) of eudicots[ref2, 3, 4]. In contrast to S-RNase, the question of whether SLF/SFB/SFBB has a single origin has been debated. Recently, Zhao et al. provided evidence through phylogenetic and functional genetic analysis that SFB, which groups with SLF and SFBB, can also neutralize the cytotoxic effects of S-RNase in hybrid petunias (P. hybrida), confirming that they share a conserved role as type 1 pollen S factors(2022). Furthermore, the SLF from the basal dicot Aquilegia coerulea also demonstrates type 1 pollen S functionality, which strengthens the case for a single origin of type 1 SI from the most recent common ancestor of eudicots[ref2].
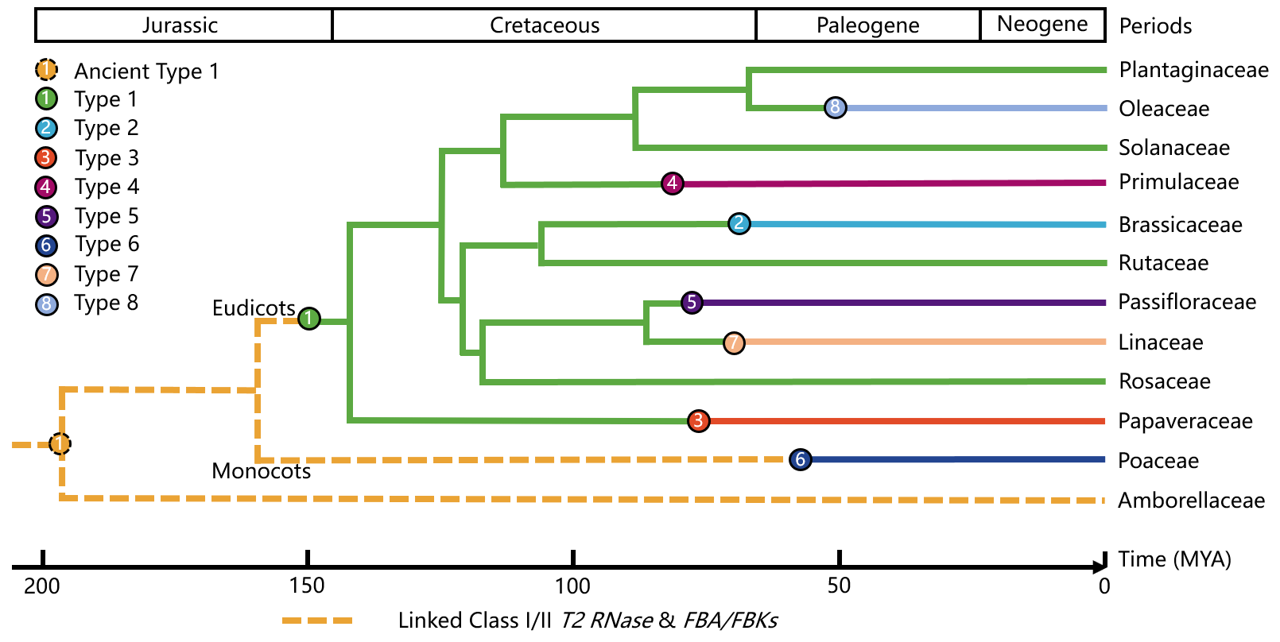
Fig. The phylogenetic tree of species at the family level illustrating the origin of the eight SI types. The evolutionary tree is generated by TimeTree. The geological time scale is shown at the top, while the bottom numerical axis represents the evolutionary timeline. The circles of different colors and their corresponding lines represent the eight types of SI. The orange dashed line indicates the ancient type-1 S structure.
During the evolution of flowering plants, the selection pressures from geographical environments and population propagation have led to frequent losses and recoveries of their self-incompatibility (SI). Zhao et al. proposed that the highly dynamic evolutionary process of SI in flowering plants mainly includes three loss approaches (I: duplication of type 1 S loci; II: inactivation of S genes; III: deletion of S loci) and four recovery approaches (a: inactivation of duplicated type 1 S loci; b: deletion of duplicated type 1 S structures; c: reactivation of S genes; d: acquisition of new S loci)[ref2]. The research found that several eudicot genomes, including the self-compatible Aquilegia and common bean (Phaseolus vulgaris), possess 2-3 type 1 S loci, indicating that the loss of type 1 SI due to whole-genome duplication and approach I is the most common. To avoid inbreeding depression, the self-incompatible Spanish snapdragon (A. hispanicum), hairy tomato (S. habrochaites), and pomelo (Citrus maxima) have recovered type 1 SI based on the inactivation or deletion of duplicated type 1 S loci. In contrast, the self-compatible cultivated snapdragon (A. majus), cultivated tomato (S. lycopersicum), and Arabidopsis (Arabidopsis thaliana) have lost SI through approach II by inactivating S-RNase and SRK/SCR, respectively. The Brassicaceae, Primulaceae, and Passifloraceae have evolved new type 2, 4, and 5 SI after completely deleting type 1 S loci, thus losing the ancient type 1 SI (approach III and d), and the Asteraceae SI may also follow this evolutionary path. Meanwhile, the Papaveraceae have evolved new type 3 S while retaining type 1 S, but the function of the former remains unclear.